Chapter 5 Parse single cell data
Seurat is a popular R package used for single-cell RNA sequencing (scRNA-seq) data analysis. It provides tools for quality control, analysis, and exploration of single-cell RNA-seq data. ClusterGvis also can visualize clustered data(different celltype clusters) with Seurat object.
We first find marker genes using Seurat::FindAllMarkers:
library(SeuratData)
library(Seurat)
data("pbmc_subset")
pbmc <- UpdateSeuratObject(object = pbmc_subset)
# find markers for every cluster compared to all remaining cells
# report only the positive ones
pbmc.markers.all <- Seurat::FindAllMarkers(pbmc,
only.pos = TRUE,
min.pct = 0.25,
logfc.threshold = 0.25)
# get top 10 genes
pbmc.markers <- pbmc.markers.all %>%
dplyr::group_by(cluster) %>%
dplyr::top_n(n = 20, wt = avg_log2FC)
# check
head(pbmc.markers)
# # A tibble: 6 × 7
# # Groups: cluster [1]
# p_val avg_log2FC pct.1 pct.2 p_val_adj cluster gene
# <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <chr>
# 1 1.96e-107 1.17 0.901 0.594 2.69e-103 Naive CD4 T LDHB
# 2 1.61e- 82 2.35 0.436 0.11 2.20e- 78 Naive CD4 T CCR7
# 3 4.20e- 77 1.08 0.838 0.406 5.76e- 73 Naive CD4 T CD3D
# 4 2.32e- 54 1.04 0.726 0.399 3.18e- 50 Naive CD4 T CD3E
# 5 3.19e- 50 1.27 0.628 0.358 4.38e- 46 Naive CD4 T NOSIP
# 6 3.32e- 49 2.09 0.336 0.104 4.56e- 45 Naive CD4 T LEF1prepareDataFromscRNA requires your Seurat object and differential expression results as input. If the showAverage parameter is set to TRUE, it means the function will calculate and plot the average expression for genes in cells of the same subgroup. If FALSE, it will plot data for all individual cells. By default, the function uses the data slot from the RNA assay in the Seurat object:
# prepare data from seurat object
st.data <- prepareDataFromscRNA(object = pbmc,
diffData = pbmc.markers,
showAverage = TRUE)
# check
str(st.data)
# List of 5
# $ wide.res:'data.frame': 177 obs. of 11 variables:
# ..$ gene : chr [1:177] "LDHB" "CCR7" "CD3D" "CD3E" ...
# ..$ Naive CD4 T : num [1:177] 1.66 2.34 1.1 1.19 1.82 ...
# ..$ Memory CD4 T: num [1:177] 1.654 0.632 1.204 1.335 1.453 ...
# ..$ CD14+ Mono : num [1:177] -0.639 -0.575 -0.649 -0.864 -0.526 ...
# ..$ B : num [1:177] -0.574 0.414 -0.671 -0.822 -0.759 ...
# ..$ CD8 T : num [1:177] 0.3047 -0.5134 1.6493 1.2945 0.0369 ...
# ..$ FCGR3A+ Mono: num [1:177] -0.886 -0.536 -0.681 -0.845 -0.144 ...
# ..$ NK : num [1:177] -0.26 -0.5575 -0.538 0.0622 -0.1026 ...
# ..$ DC : num [1:177] -0.521 -0.53 -0.685 -0.865 -0.63 ...
# ..$ Platelet : num [1:177] -0.74 -0.679 -0.728 -0.488 -1.143 ...
# ..$ cluster : chr [1:177] "1" "1" "1" "1" ...
# $ long.res:'data.frame': 1593 obs. of 5 variables:
# ..$ cluster : chr [1:1593] "1" "1" "1" "1" ...
# ..$ gene : chr [1:1593] "LDHB" "CCR7" "CD3D" "CD3E" ...
# ..$ cell_type : Factor w/ 9 levels "Naive CD4 T",..: 1 1 1 1 1 1 1 1 1 1 ...
# ..$ norm_value : num [1:1593] 1.66 2.34 1.1 1.19 1.82 ...
# ..$ cluster_name: Factor w/ 9 levels "cluster 1 (20)",..: 1 1 1 1 1 1 1 1 1 1 ...
# $ type : chr "scRNAdata"
# $ geneMode: chr "average"
# $ geneType: chr "unique|_"Enrichment anaysis for each celltype:
library(org.Hs.eg.db)
# enrich for clusters
enrich <- enrichCluster(object = st.data,
OrgDb = org.Hs.eg.db,
type = "BP",
organism = "hsa",
pvalueCutoff = 0.5,
topn = 5)
# check
head(enrich)
# group Description pvalue ratio
# GO:0030098 C1 lymphocyte differentiation 4.255042e-09 42.10526
# GO:1903131...2 C1 mononuclear cell differentiation 1.034991e-08 42.10526
# GO:0030217 C1 T cell differentiation 1.275815e-08 36.84211
# GO:0050863...4 C1 regulation of T cell activation 5.354207e-08 36.84211
# GO:0022407 C1 regulation of cell-cell adhesion 3.163499e-07 36.84211
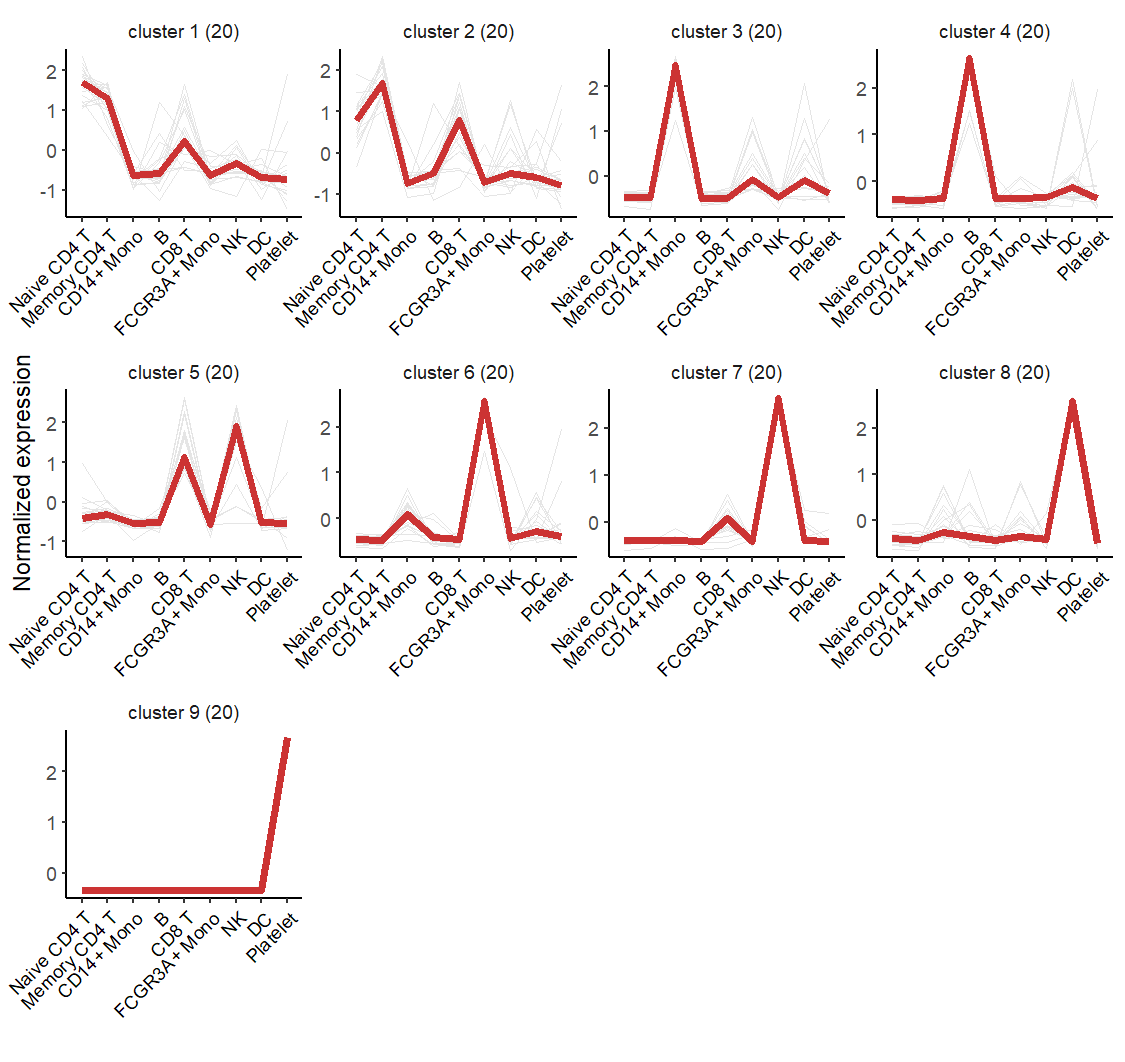
# GO:0002260 C2 lymphocyte homeostasis 4.246017e-07 25.00000Line plot:
# add gene name
markGenes = unique(pbmc.markers$gene)[sample(1:length(unique(pbmc.markers$gene)),40,
replace = F)]
# line plot
visCluster(object = st.data,
plot.type = "line")
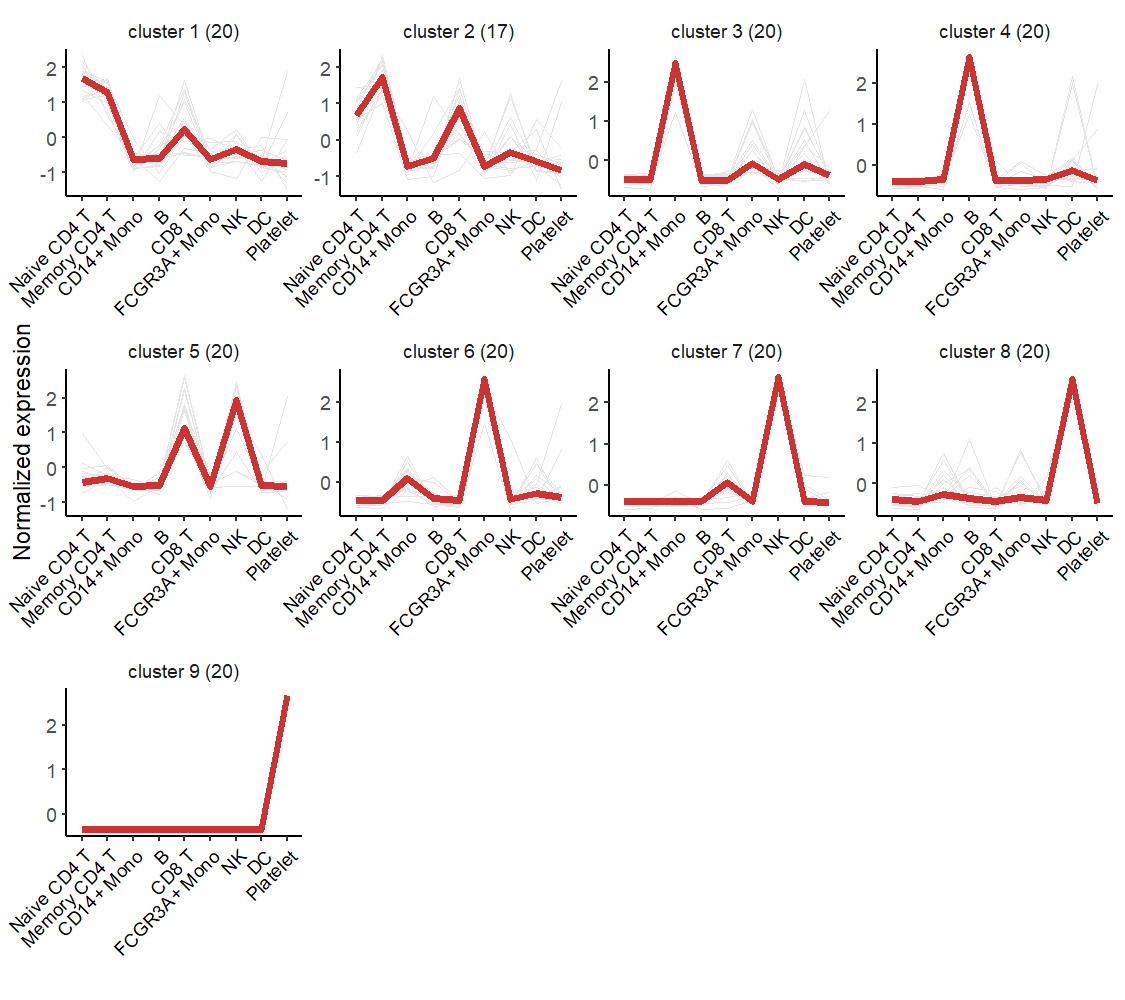
Order the cluster:
# heatmap plot
pdf('sc1.pdf',height = 10,width = 6,onefile = F)
visCluster(object = st.data,
plotType = "heatmap",
column_names_rot = 45,
markGenes = markGenes,
clusterOrder = c(1:9))
dev.off()
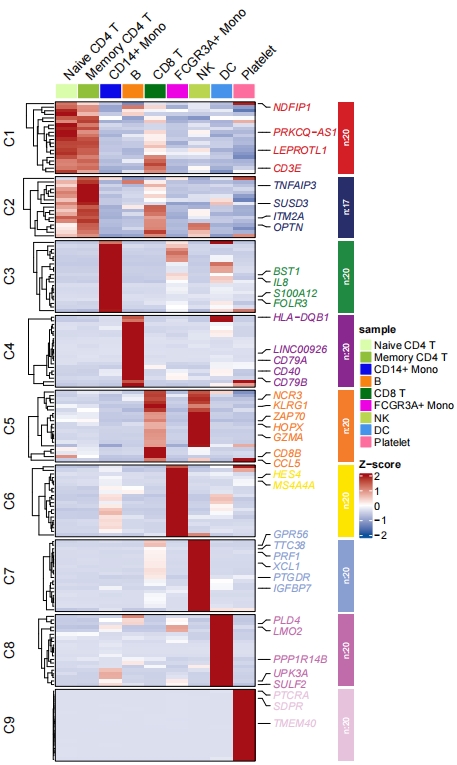
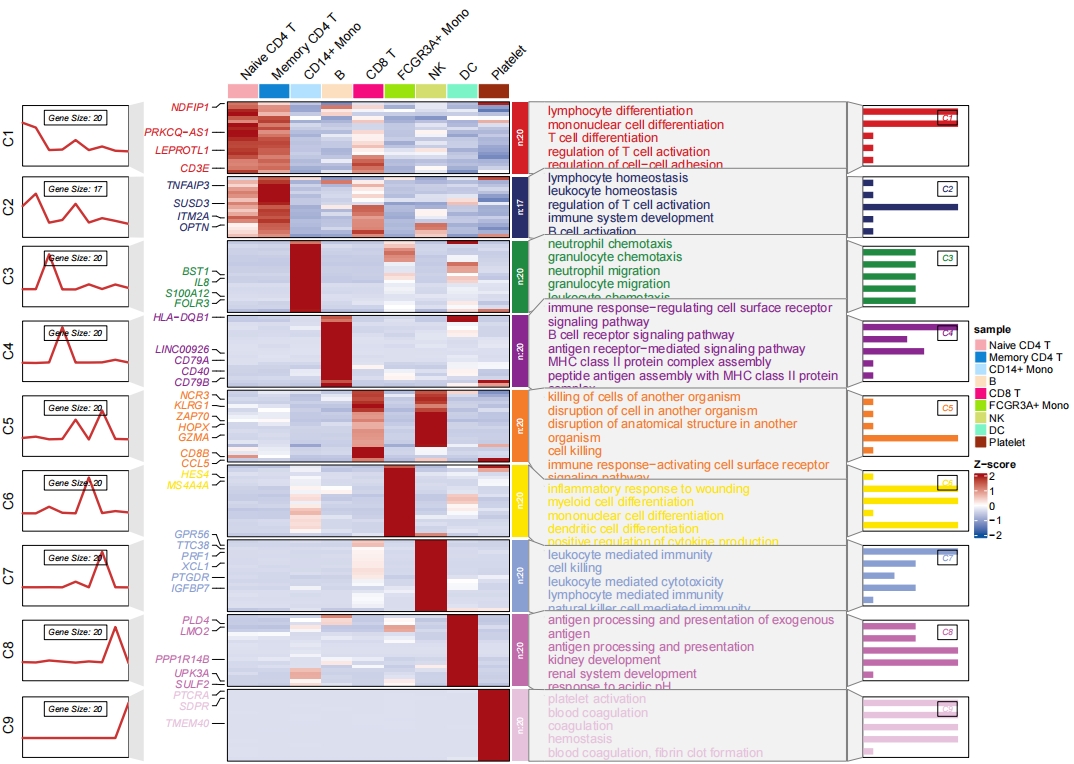
Add enrichment annotation:
# add bar plot
pdf('sc2.pdf',height = 10,width = 14,onefile = F)
visCluster(object = st.data,
plotType = "both",
column_names_rot = 45,
show_row_dend = F,
markGenes = markGenes,
markGenesSide = "left",
annoTermData = enrich,
lineSide = "left",
clusterOrder = c(1:9),
goCol = rep(jjAnno::useMyCol("stallion",n = 9),each = 5),
addBar = T)
dev.off()
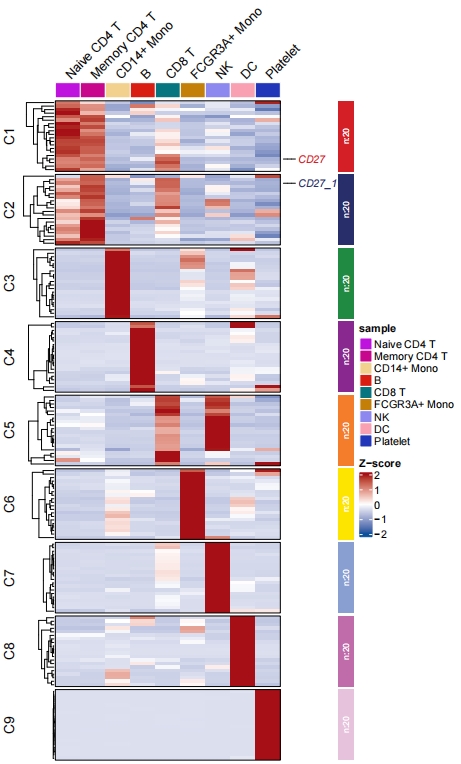
During differential expression analysis, there may be multiple identical genes that show differential expression across several subgroups simultaneously. By default, these duplicate genes will be deduplicated. This might result in fewer genes available for enrichment analysis. However, you can use the parameter keep.uniqGene=FALSE to retain duplicate genes:
# retain duplicate diff gene in multiple clusters
st.data <- prepareDataFromscRNA(object = pbmc,
diffData = pbmc.markers,
showAverage = TRUE,
keepUniqGene = FALSE,
sep = "_")
# check
df <- st.data$wide.res
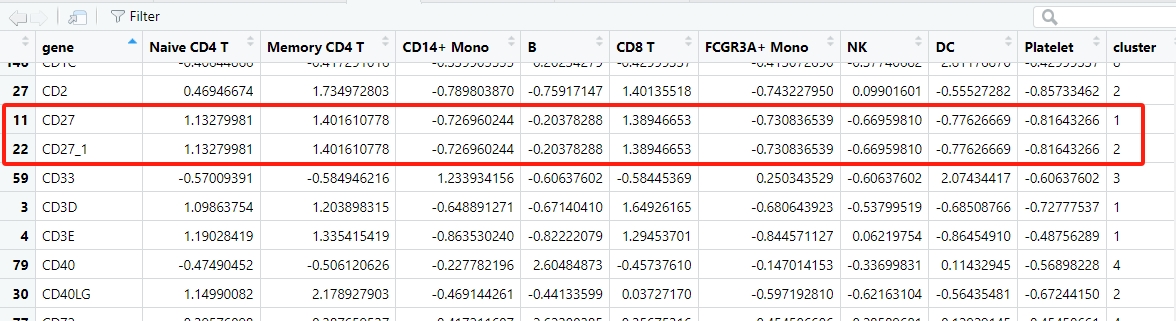
Line plot without de-duplicate genes:
Let’s see the duplicate genes in heatmap:
# heatmap plot
pdf('sc3.pdf',height = 10,width = 6,onefile = F)
visCluster(object = st.data,
plotType = "heatmap",
column_names_rot = 45,
markGenes = c("CD27","CD27_1"),
clusterOrder = c(1:9))
dev.off()
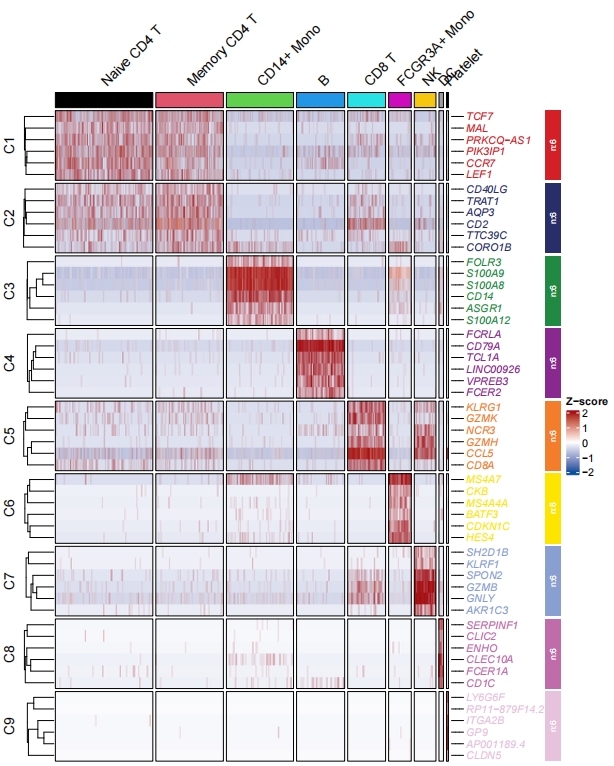
Now let’s show all cells expression:
# no average cells
pbmc.markers1 <- pbmc.markers.all %>%
dplyr::group_by(cluster) %>%
dplyr::top_n(n = 6, wt = avg_log2FC)
# retain duplicate diff gene in multiple clusters
st.data <- prepareDataFromscRNA(object = pbmc,
diffData = pbmc.markers1,
showAverage = FALSE)
# check
str(st.data)
# List of 5
# $ wide.res:'data.frame': 54 obs. of 2640 variables:
# ..$ gene : chr [1:54] "MAL" "LEF1" "TCF7" "PRKCQ-AS1" ...
# ..$ AAACGCTGTAGCCA|Naive CD4 T : num [1:54] -0.381 -0.434 2.272 -0.439 -0.479 ...
# ..$ AAACTTGATCCAGA|Naive CD4 T : num [1:54] -0.381 -0.434 1.585 -0.439 2.914 ...
# ..$ AAAGAGACGAGATA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 1.927 -0.479 ...
# ..$ AAAGAGACGGACTT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AAAGAGACGGCATT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 3.332 -0.479 ...
# ..$ AAAGTTTGTAGAGA|Naive CD4 T : num [1:54] -0.381 -0.434 2.44 -0.439 2.527 ...
# ..$ AAATCAACACCAGT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 3.486 -0.479 ...
# ..$ AAATCAACCAGGAG|Naive CD4 T : num [1:54] 2.01 1.78 1.42 1.76 2.25 ...
# ..$ AAATCAACGGAAGC|Naive CD4 T : num [1:54] -0.381 2.353 1.211 1.518 -0.479 ...
# ..$ AAATCCCTCCACAA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AAATTCGAAGGTTC|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 1.521 ...
# ..$ AAATTCGAGGAGTG|Naive CD4 T : num [1:54] -0.381 1.713 -0.534 -0.439 1.436 ...
# ..$ AACAAACTCATTTC|Naive CD4 T : num [1:54] 2.32 2.08 2.45 2.05 1.76 ...
# ..$ AACAAACTTTCGTT|Naive CD4 T : num [1:54] 2.058 1.829 -0.534 -0.439 1.54 ...
# ..$ AACAATACGACGAG|Naive CD4 T : num [1:54] -0.381 2.118 1.718 -0.439 2.581 ...
# ..$ AACACGTGGAAAGT|Naive CD4 T : num [1:54] -0.381 -0.434 2.237 -0.439 -0.479 ...
# ..$ AACACGTGGCTACA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 1.817 -0.479 ...
# ..$ AACCCAGATCGCTC|Naive CD4 T : num [1:54] 2.231 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AACCGATGCTCCCA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 1.985 1.705 ...
# ..$ AACCTACTGTGTTG|Naive CD4 T : num [1:54] 2.064 -0.434 1.469 1.807 2.302 ...
# ..$ AACCTTACGAGACG|Naive CD4 T : num [1:54] -0.381 2.638 1.433 1.767 1.509 ...
# ..$ AACCTTTGGACGGA|Naive CD4 T : num [1:54] -0.381 -0.434 1.733 -0.439 -0.479 ...
# ..$ AACGCATGACCCAA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AACGCATGCCTTCG|Naive CD4 T : num [1:54] -0.381 1.756 -0.534 -0.439 1.475 ...
# ..$ AACGCATGTACTTC|Naive CD4 T : num [1:54] -0.381 -0.434 1.383 -0.439 -0.479 ...
# ..$ AACGTGTGTCCAAG|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 2.371 -0.479 ...
# ..$ AACTACCTTAGAGA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 3.038 ...
# ..$ AACTTGCTGGGACA|Naive CD4 T : num [1:54] -0.381 2.265 1.849 -0.439 -0.479 ...
# ..$ AAGAACGAGTGTTG|Naive CD4 T : num [1:54] 3.747 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AAGACAGAGGATCT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 1.974 -0.479 ...
# ..$ AAGATTACAACCTG|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 2.08 -0.479 ...
# ..$ AAGCACTGCATACG|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AAGCCAACGTGTTG|Naive CD4 T : num [1:54] -0.381 -0.434 1.436 -0.439 1.512 ...
# ..$ AAGCCATGCGTGAT|Naive CD4 T : num [1:54] -0.381 2.114 1.715 -0.439 3.064 ...
# ..$ AAGCCATGTCTCGC|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AAGCGACTGTGTCA|Naive CD4 T : num [1:54] -0.381 2.482 2.04 1.631 2.123 ...
# ..$ AAGCGTACGTCTTT|Naive CD4 T : num [1:54] -0.381 2.151 1.748 -0.439 2.614 ...
# ..$ AAGGTCTGCAGATC|Naive CD4 T : num [1:54] -0.381 -0.434 2.592 -0.439 2.681 ...
# ..$ AAGTATACCGAACT|Naive CD4 T : num [1:54] -0.381 -0.434 1.999 -0.439 -0.479 ...
# ..$ AAGTCCGACTTGTT|Naive CD4 T : num [1:54] -0.381 2.517 -0.534 -0.439 2.154 ...
# ..$ AAGTCCGATAGAAG|Naive CD4 T : num [1:54] -0.381 1.576 -0.534 -0.439 -0.479 ...
# ..$ AAGTCTCTAGTCGT|Naive CD4 T : num [1:54] -0.381 1.605 2.448 -0.439 -0.479 ...
# ..$ AATAAGCTCGTTGA|Naive CD4 T : num [1:54] -0.381 3.358 2.013 -0.439 2.096 ...
# ..$ AATCCTACCGGTAT|Naive CD4 T : num [1:54] 2.179 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AATCGGTGTGCTTT|Naive CD4 T : num [1:54] -0.381 2.245 -0.534 -0.439 3.55 ...
# ..$ AATCTAGAATCGGT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ AATCTCACTCTAGG|Naive CD4 T : num [1:54] -0.381 2.093 1.697 -0.439 -0.479 ...
# ..$ AATCTCTGAACAGA|Naive CD4 T : num [1:54] -0.381 2.073 2.451 2.908 1.758 ...
# ..$ AATGGCTGTGAAGA|Naive CD4 T : num [1:54] -0.381 -0.434 1.699 2.066 -0.479 ...
# ..$ AATGTAACGGTGGA|Naive CD4 T : num [1:54] 3.366 1.687 -0.534 -0.439 2.154 ...
# ..$ AATTACGACTTCTA|Naive CD4 T : num [1:54] -0.381 -0.434 1.51 3.228 -0.479 ...
# ..$ ACAAAGGAGGGTGA|Naive CD4 T : num [1:54] -0.381 2.169 1.763 -0.439 -0.479 ...
# ..$ ACAAGAGACTTATC|Naive CD4 T : num [1:54] -0.381 -0.434 2.617 -0.439 -0.479 ...
# ..$ ACAGGTACCCCACT|Naive CD4 T : num [1:54] 2.04 -0.434 1.449 -0.439 1.526 ...
# ..$ ACAGTCGACCCAAA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ ACAGTGTGGTCACA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ ACATGGTGAAGCCT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ ACATGGTGCGAGTT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 2.993 -0.479 ...
# ..$ ACATTCTGGCATAC|Naive CD4 T : num [1:54] -0.381 1.763 1.405 -0.439 -0.479 ...
# ..$ ACCAACGACATGCA|Naive CD4 T : num [1:54] -0.381 -0.434 1.713 -0.439 -0.479 ...
# ..$ ACCACAGAAAGTAG|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ ACCACAGAGTTGGT|Naive CD4 T : num [1:54] -0.381 -0.434 1.403 2.563 -0.479 ...
# ..$ ACCACCTGTGTGCA|Naive CD4 T : num [1:54] -0.381 2.85 -0.534 2.812 2.451 ...
# ..$ ACCACGCTACAGCT|Naive CD4 T : num [1:54] 2.134 1.899 1.526 -0.439 2.366 ...
# ..$ ACCACGCTGCGAGA|Naive CD4 T : num [1:54] -0.381 2.192 -0.534 -0.439 -0.479 ...
# ..$ ACCACGCTGCTGTA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ ACCAGTGAGGGATG|Naive CD4 T : num [1:54] 2.177 2.8 -0.534 1.911 -0.479 ...
# ..$ ACCATTACCTTCTA|Naive CD4 T : num [1:54] 1.77 1.56 1.23 1.54 1.3 ...
# ..$ ACCATTTGTCATTC|Naive CD4 T : num [1:54] -0.381 2.419 -0.534 -0.439 2.066 ...
# ..$ ACCCACTGGACAGG|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 2.817 ...
# ..$ ACCCAGCTTGCTTT|Naive CD4 T : num [1:54] 2.151 -0.434 2.297 1.887 -0.479 ...
# ..$ ACCCGTTGCTGCAA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 2.057 1.771 ...
# ..$ ACCTCCGATATGCG|Naive CD4 T : num [1:54] 2.583 -0.434 -0.534 -0.439 -0.479 ...
# ..$ ACCTCCGATGCTGA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ ACGAAGCTCTCCAC|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 3.327 -0.479 ...
# ..$ ACGACCCTGATGAA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 2.141 -0.479 ...
# ..$ ACGACCCTTGACCA|Naive CD4 T : num [1:54] -0.381 -0.434 1.618 -0.439 -0.479 ...
# ..$ ACGAGGGACGAACT|Naive CD4 T : num [1:54] 3.927 1.8 -0.534 1.772 -0.479 ...
# ..$ ACGATTCTACGGGA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 2.416 -0.479 ...
# ..$ ACGCCGGAAAGCCT|Naive CD4 T : num [1:54] -0.381 2.096 -0.534 -0.439 1.778 ...
# ..$ ACGCTCACAGTACC|Naive CD4 T : num [1:54] 2.218 -0.434 1.594 -0.439 1.672 ...
# ..$ ACGCTCACCCTTGC|Naive CD4 T : num [1:54] 3.277 -0.434 1.69 2.055 1.769 ...
# ..$ ACGGCTCTTGCACA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 1.868 -0.479 ...
# ..$ ACGGTAACGGTGGA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 2.893 ...
# ..$ ACGGTCCTCGGGAA|Naive CD4 T : num [1:54] -0.381 2.035 -0.534 -0.439 -0.479 ...
# ..$ ACGTCAGAGGGATG|Naive CD4 T : num [1:54] -0.381 2.502 -0.534 -0.439 -0.479 ...
# ..$ ACGTGATGGGTCTA|Naive CD4 T : num [1:54] 2.084 -0.434 1.485 2.667 -0.479 ...
# ..$ ACGTTACTTTCCAT|Naive CD4 T : num [1:54] -0.381 -0.434 2.572 -0.439 1.871 ...
# ..$ ACGTTGGAAAAGCA|Naive CD4 T : num [1:54] 3.261 -0.434 2.449 -0.439 1.756 ...
# ..$ ACGTTTACATCAGC|Naive CD4 T : num [1:54] -0.381 2.841 2.357 1.948 -0.479 ...
# ..$ ACTAAAACCCACAA|Naive CD4 T : num [1:54] -0.381 -0.434 2.279 -0.439 -0.479 ...
# ..$ ACTACGGACCTATT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 1.965 ...
# ..$ ACTACGGATCGCTC|Naive CD4 T : num [1:54] -0.381 -0.434 2.782 1.898 1.627 ...
# ..$ ACTACTACTAAGGA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# ..$ ACTATCACTGCCAA|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 2.517 ...
# ..$ ACTCAGGACTGAAC|Naive CD4 T : num [1:54] 3.279 2.962 1.691 -0.439 3.036 ...
# ..$ ACTCTCCTGACACT|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 3.222 ...
# ..$ ACTCTCCTGCATAC|Naive CD4 T : num [1:54] -0.381 -0.434 -0.534 -0.439 -0.479 ...
# .. [list output truncated]
# $ long.res:'data.frame': 142452 obs. of 5 variables:
# ..$ cluster : chr [1:142452] "1" "1" "1" "1" ...
# ..$ gene : chr [1:142452] "MAL" "LEF1" "TCF7" "PRKCQ-AS1" ...
# ..$ cell_type : chr [1:142452] "Naive CD4 T" "Naive CD4 T" "Naive CD4 T" "Naive CD4 T" ...
# ..$ norm_value : num [1:142452] -0.381 -0.434 2.272 -0.439 -0.479 ...
# ..$ cluster_name: Factor w/ 9 levels "cluster 1 (6)",..: 1 1 1 1 1 1 1 1 1 1 ...
# $ type : chr "scRNAdata"
# $ geneMode: chr "all"
# $ geneType: chr "unique|_"Plot:
# heatmap plot
pdf('sc4.pdf',height = 10,width = 8,onefile = F)
visCluster(object = st.data,
plotType = "heatmap",
markGenes = unique(pbmc.markers1$gene),
column_title_rot = 45,
clusterOrder = 1:9)
dev.off()
Add enrichment annotation:
# add GO annotation
pdf('sc6.pdf',height = 12,width = 16,onefile = F)
visCluster(object = st.data,
plotType = "both",
column_title_rot = 45,
markGenes = unique(pbmc.markers1$gene),
markGenesSide = "left",
annoTermData = enrich,
lineSide = "left",
clusterOrder = c(1:9),
addBar = T)
dev.off()
