Chapter 10 Createing your own genome track
You can supply your own genome chromosome size data to create your own genome track besides the model organisms. Here is a simple example.
my_chr <- data.frame(chr = paste0("Chr",1:4),size = sample(seq(1,100,1),4,replace = T))
my_chr
# chr size
# 1 Chr1 98
# 2 Chr2 18
# 3 Chr3 84
# 4 Chr4 5
chr_df <- data.frame(chr = my_chr$chr,start = 0,end = my_chr$size)
chr_df
# chr start end
# 1 Chr1 0 98
# 2 Chr2 0 18
# 3 Chr3 0 84
# 4 Chr4 0 5
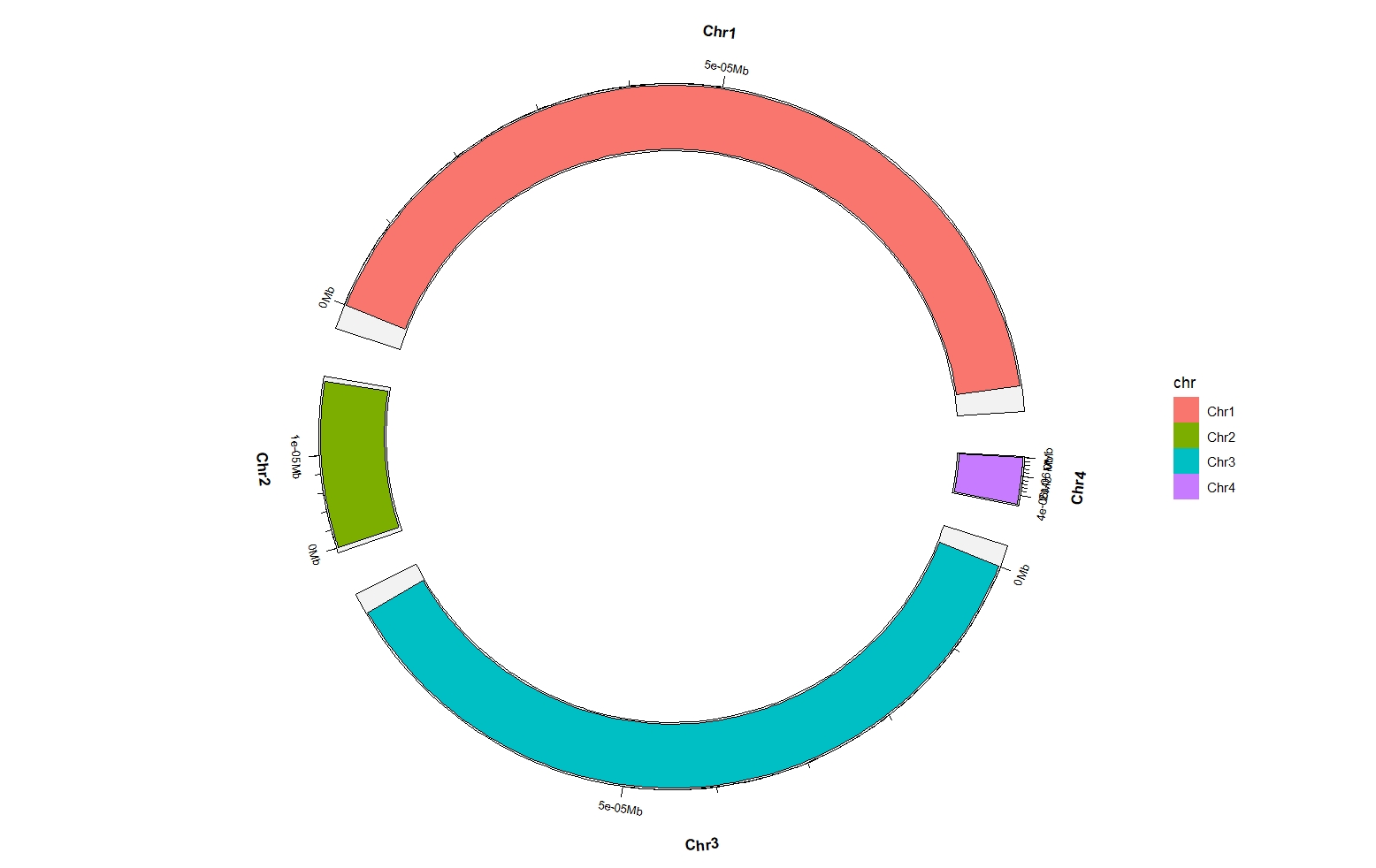
ggcirclize(chr_df,
aes(end = 360,genome = "my_chr",
chr = chr,gstart = start,gend = end)) +
ggcirclize::geom_trackgenomicrect(aes(r0 = 0.8,r1 = 1,fill = chr))