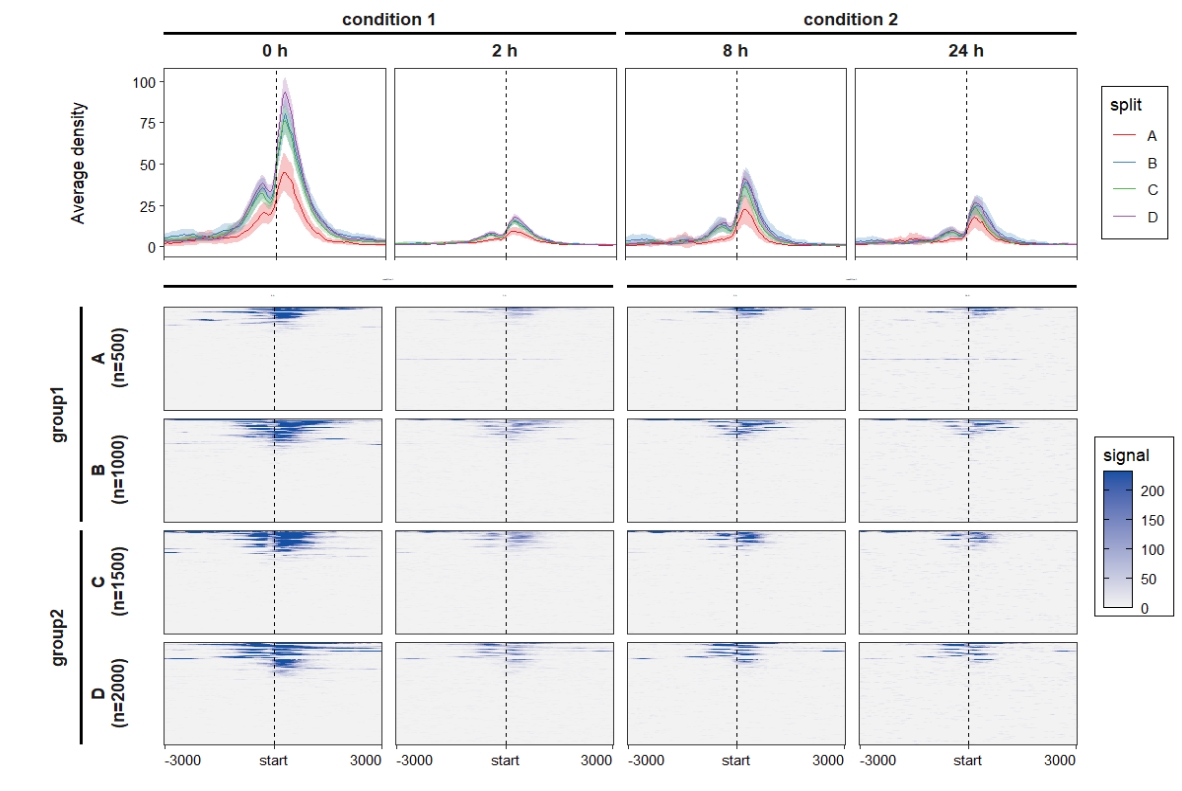
Chapter 5 Adding rowsplit groups
Imaging you have multiple groups of binding sites, we can also group them into multiple subset panels in plot. The examples showed here.
5.1 Prepare data
Here we give a list to group.split to define how the rowsplits are grouped by:
library(ggChIPvis)
library(ggplot2)
# parse deeptools output
deep_mat <- parseDeeptools(deeptools_output = "refPoint-data.gz")
# add rowsplit groups
mat_df <- retriveData(mat.list = deep_mat,rm.extreme.value = T,
sample.names = c("0 h","2 h","8 h","24 h"),
group.sample = rep(c("condition 1","condition 2"),each = 2),
row.split = rep(c(LETTERS[1:4]),c(500,1000,1500,2000)),
group.split = list(group1 = c("A","B"),
group2 = c("C","D")))5.2 Visualization data
He we show the heatmap plot:
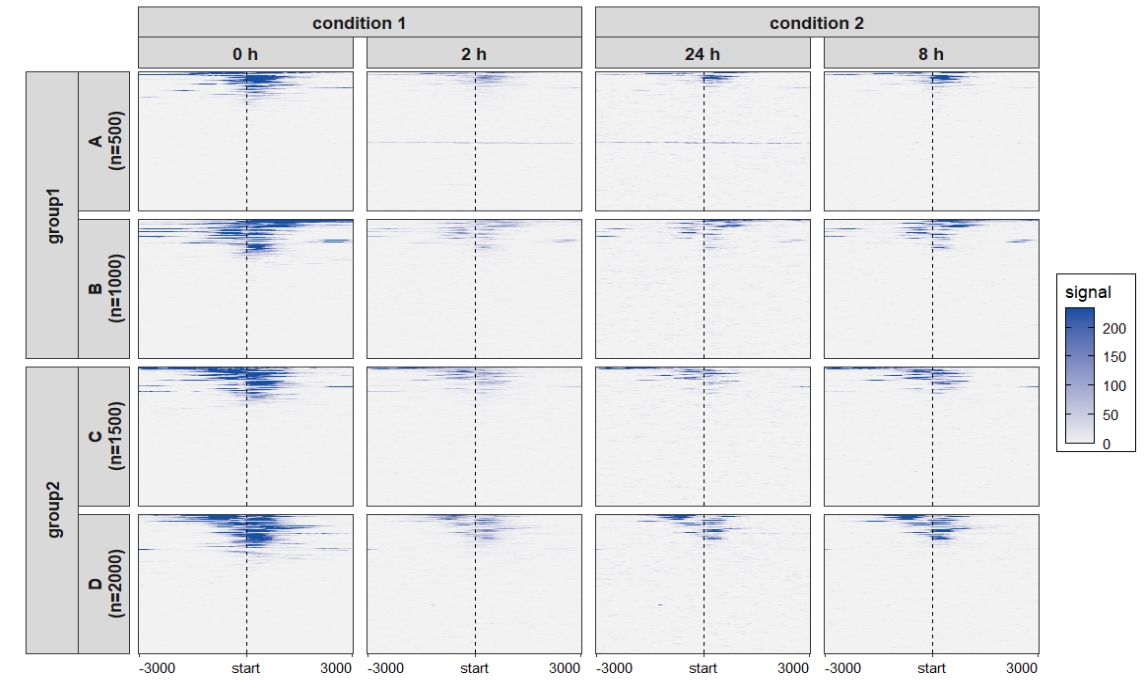 Add nested line:
Add nested line:
ChipVis(object = mat_df,plot.type = "heatmap",
sample.order = c("0 h","2 h","8 h","24 h"),
theme_params = list(strip.background = element_blank(),
ggh4x.facet.nestline = element_line(linewidth = 1,
colour = "black")))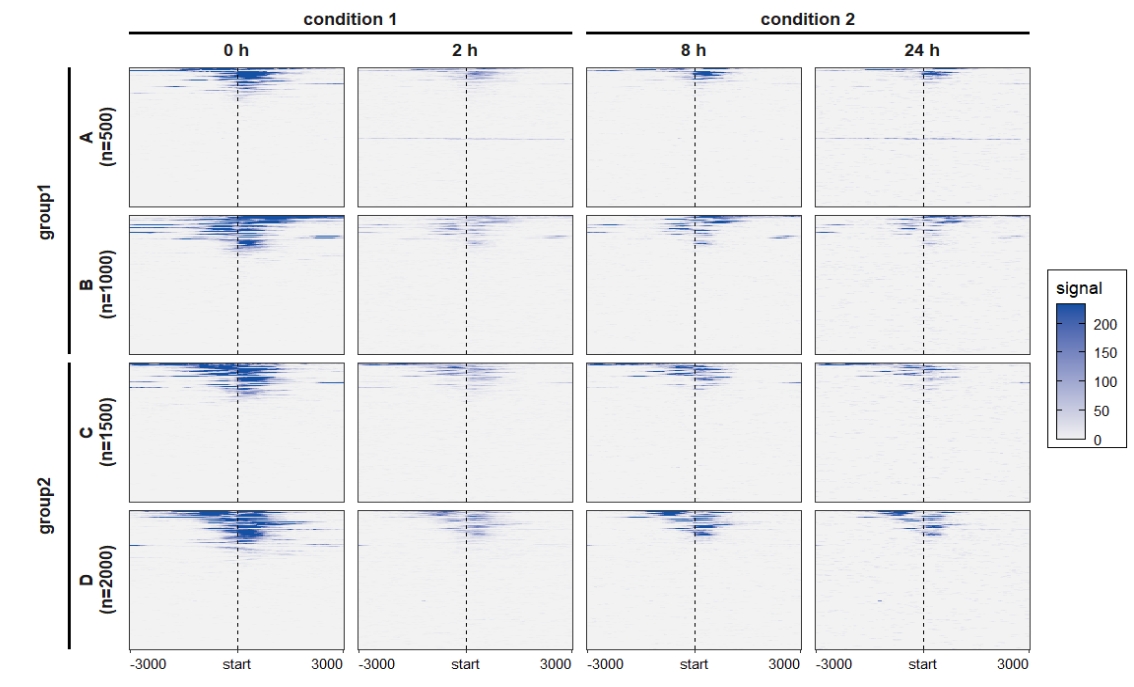 Change the rowsplit groups order:
Change the rowsplit groups order:
ChipVis(object = mat_df,plot.type = "heatmap",
sample.order = c("0 h","2 h","8 h","24 h"),
theme_params = list(strip.background = element_blank(),
ggh4x.facet.nestline = element_line(linewidth = 1,
colour = "black")),
rowgroup.order = c("group2","group1"))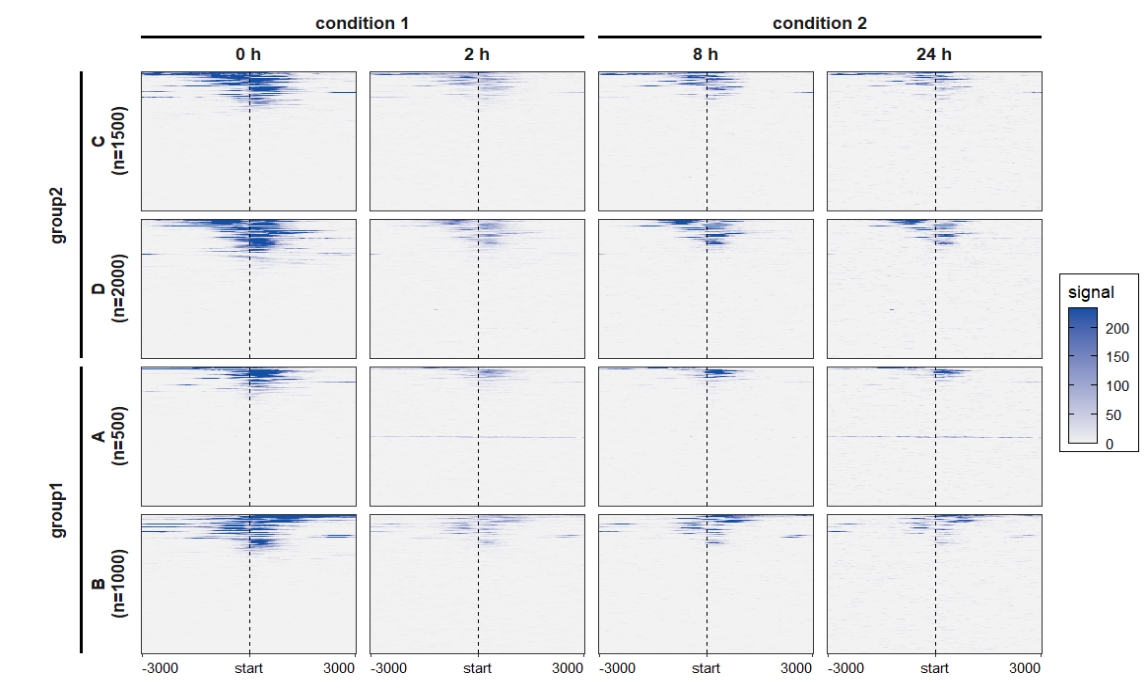
Combine the profile and heatmap plot:
ChipVis(object = mat_df,plot.type = "both",
sample.order = c("0 h","2 h","8 h","24 h"),
facet_profile_params = list(nest_line = element_line(linetype = 1)),
add_nested_line = T)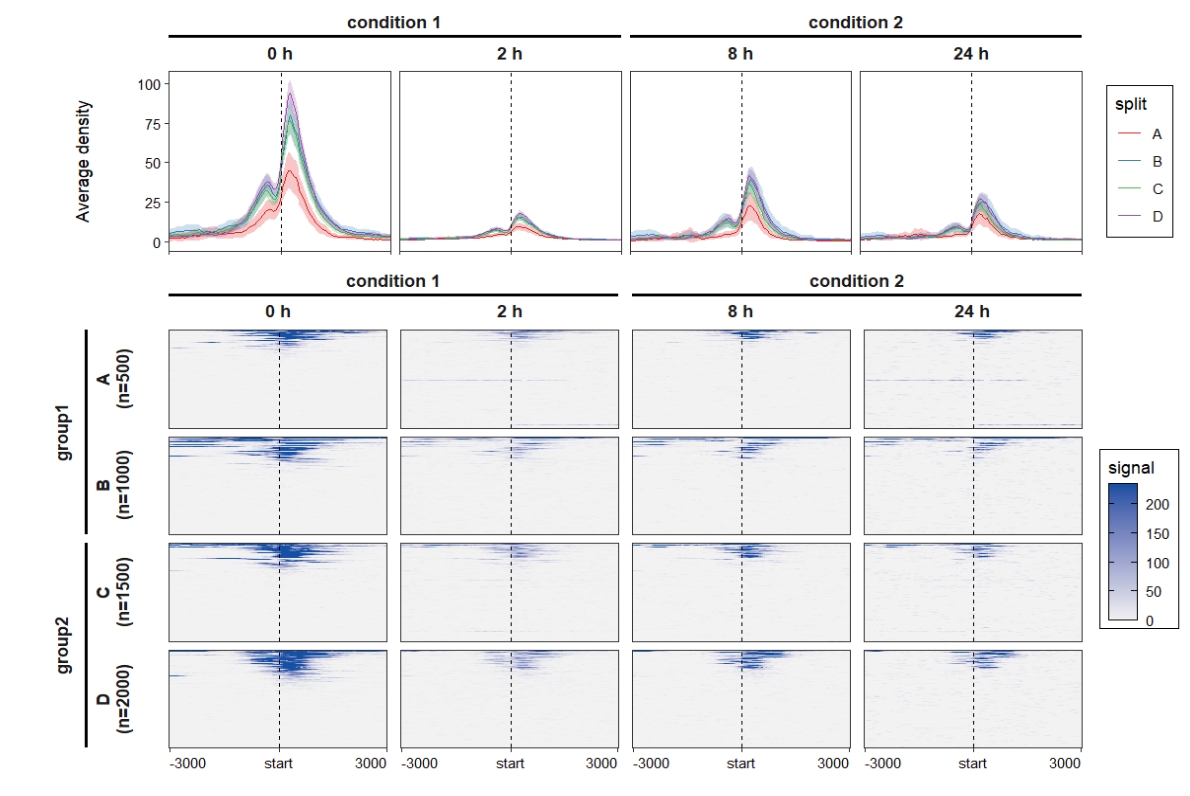 Remove the upper text of the heatmap:
Remove the upper text of the heatmap:
ChipVis(object = mat_df,plot.type = "both",
sample.order = c("0 h","2 h","8 h","24 h"),
facet_profile_params = list(nest_line = element_line(linetype = 1)),
facet_heatmap_params = list(strip = ggh4x::strip_nested(text_x = ggh4x::elem_list_text(size = 0)),
nest_line = element_line(linetype = 1)),
add_nested_line = T)