Chapter 6 Statistics of enriched signals
We can use the extracted data to draw any plot what you want. One example is that we can do some statistics for the enrichment signals across the samples.
6.2 Drawing boxplot
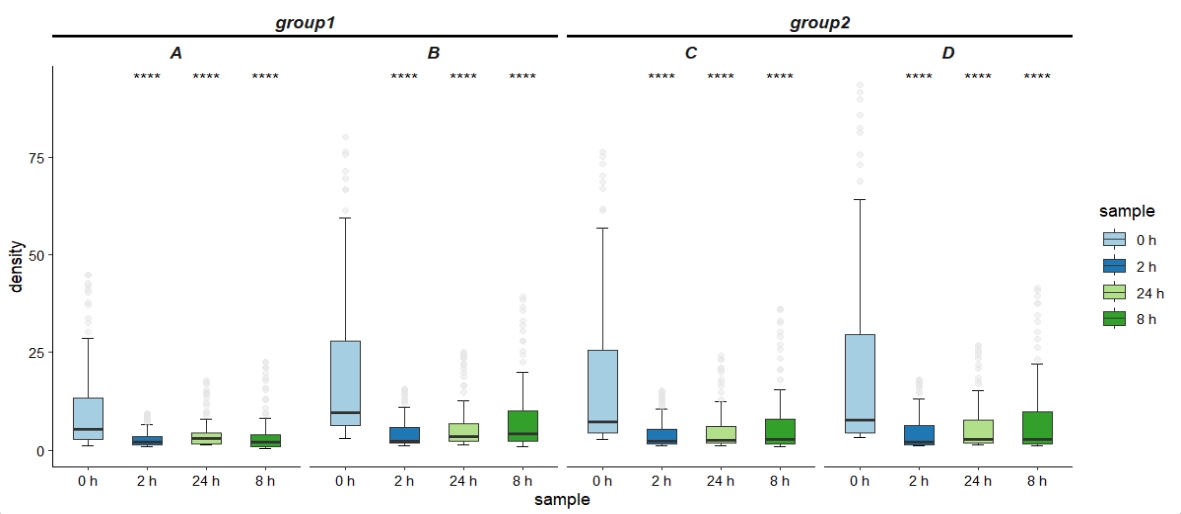
We can see the difference across different time point on multiple binding-site groups:
library(ggpubr)
library(ggh4x)
# plot
ggplot(df.profile.data,
aes(x = sample,y = density,fill = sample)) +
stat_boxplot(aes(ymin = ..lower.., ymax = ..upper..),outlier.shape = NA,width=0.5) +
stat_boxplot(geom = "errorbar", aes(ymin = ..ymax..),width=0.2,size=0.35) +
stat_boxplot(geom = "errorbar", aes(ymax = ..ymin..),width=0.2,size=0.35) +
geom_boxplot(outlier.color = "grey90",outlier.alpha = 0.5,
width=0.5,alpha = 0.5) +
facet_nested(~split.group + split,
nest_line = element_line(linetype = 1)) +
theme_classic() +
scale_fill_brewer(palette = "Paired") +
theme(strip.background = element_blank(),
axis.text = element_text(colour = "black"),
strip.text = element_text(face = "bold.italic",size = rel(1)),
ggh4x.facet.nestline = element_line(linewidth = 1,
colour = "black")) +
stat_compare_means(ref.group = "0 h",label = "p.signif")