Chapter 8 multiHeatmap based on grid
multiHeatmap is mainly based on ChipHeatmap to produce plot and parameters can be passed with it.
8.1 Combine multiple plot
multiHeatmap need accept normalizeToMatrix/getTagMatrix/parseDeeptools output:
col2 <- RColorBrewer::brewer.pal(4,"Set1")
col <- list(c("white",col2[1]),c("white",col2[2]),
c("white",col2[3]),c("white",col2[4]))
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"))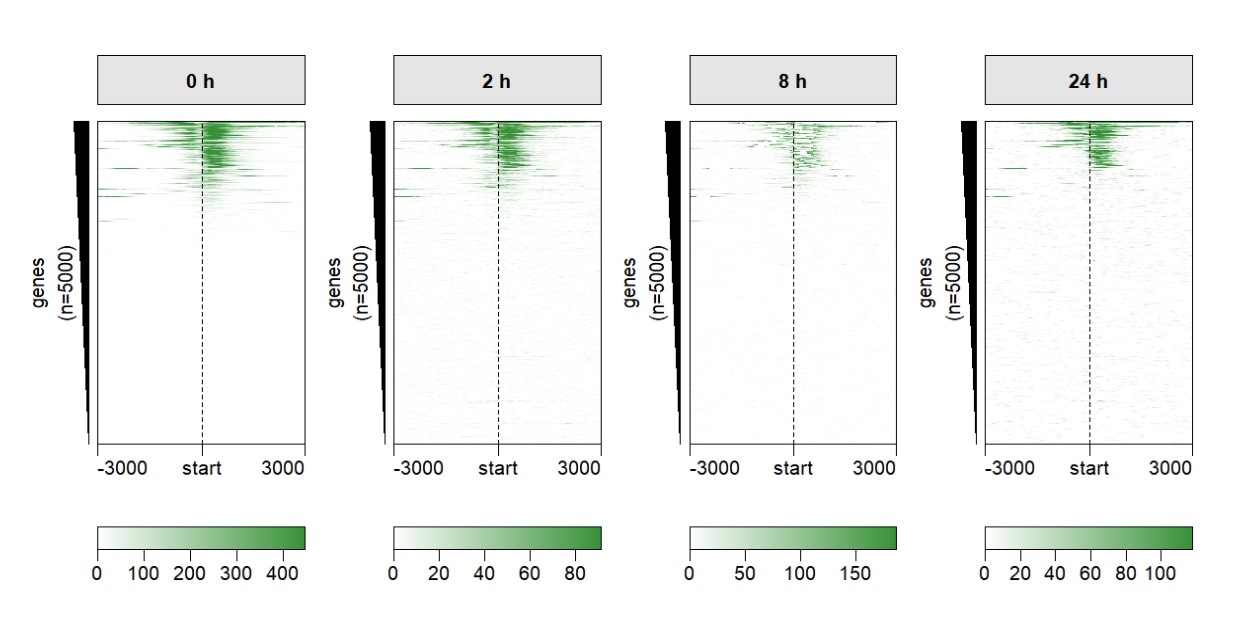 Control sample label style:
Control sample label style:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
ChipHeatmap.params = list(label.rect.gp = gpar(fill = NA,col = NA)))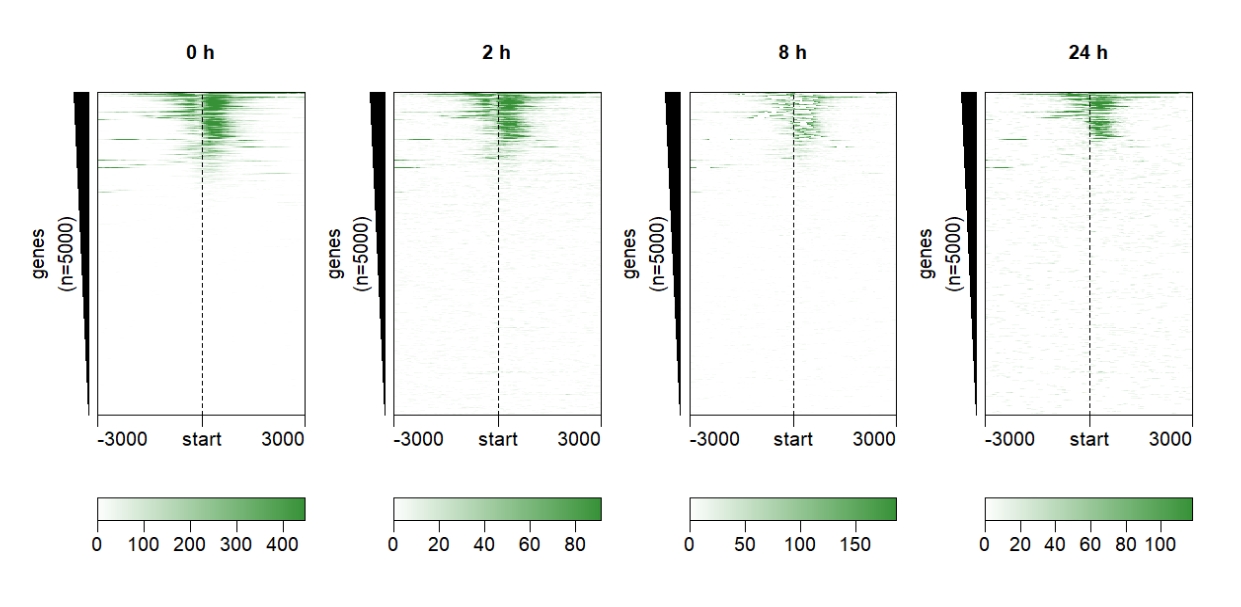 Change the heatmap color:
Change the heatmap color:
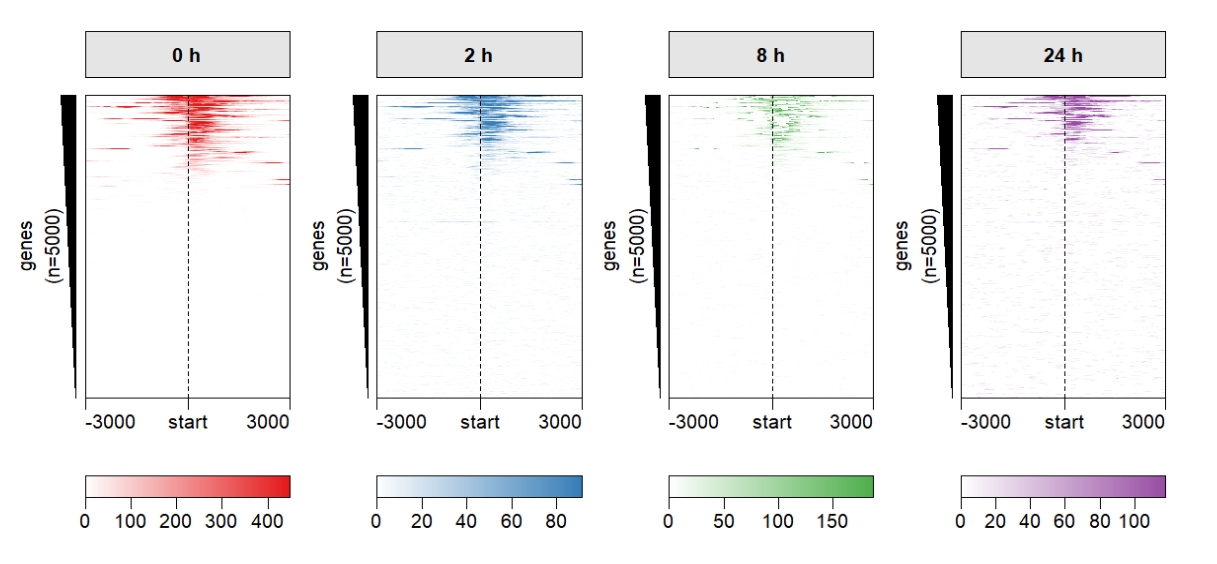 Keep one left annobar if your row orders and numbers are same:
Keep one left annobar if your row orders and numbers are same:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
heatmap.col = col,
keep.one.left.annobar = T)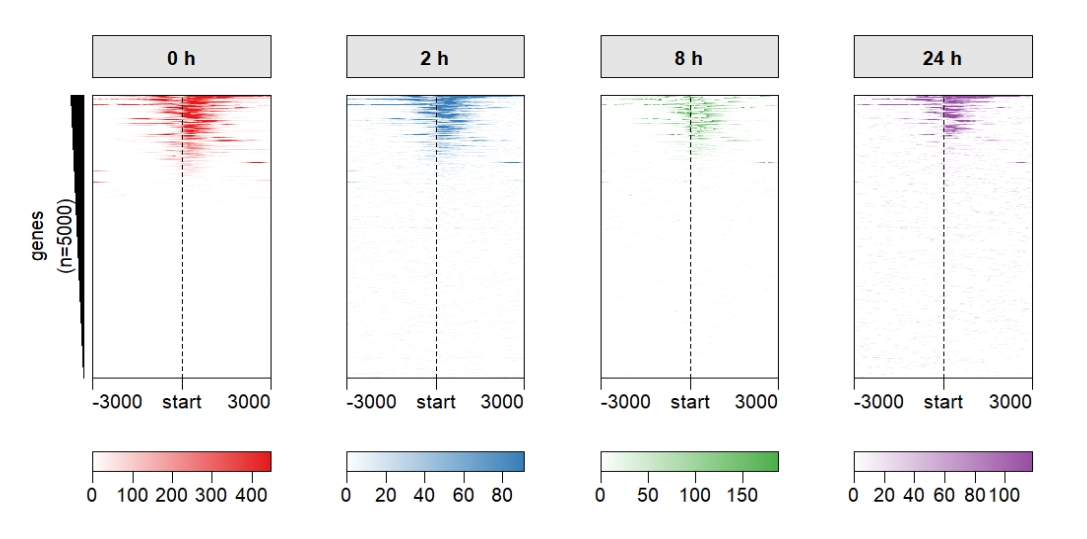 Supply with ChipHeatmap parameters:
Supply with ChipHeatmap parameters:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
heatmap.col = col,
keep.one.left.annobar = T,
ChipHeatmap.params = list(HeatmapLayout.params = list(heatmap.size = c(0.9,0.5)),
ht.xaxis.rot = 45))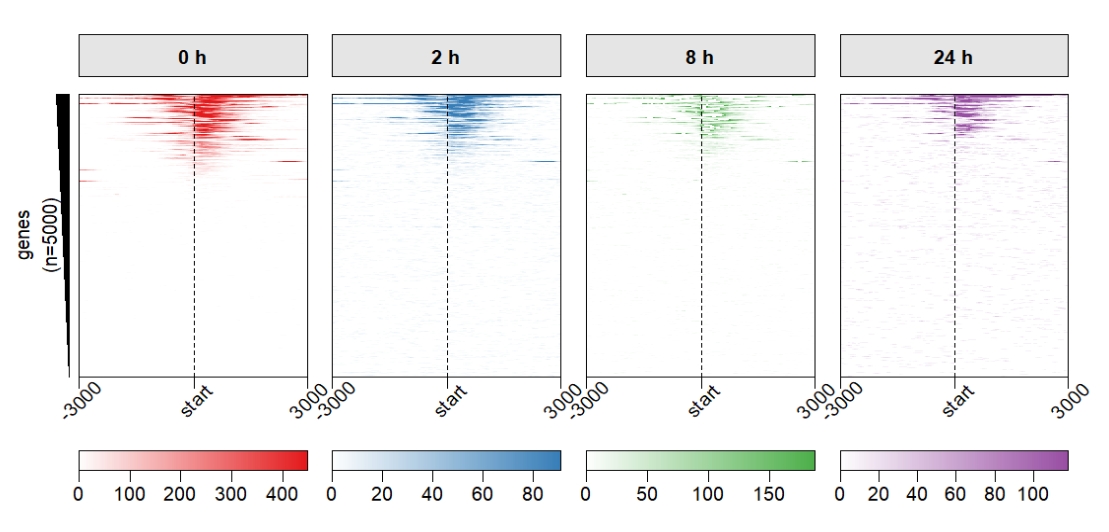
scale.range can make the multiple legend signal range to be same, so you can keep just one legend:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
keep.one.left.annobar = T,
ChipHeatmap.params = list(HeatmapLayout.params = list(heatmap.size = c(0.9,0.5))),
scale.range = T,
keep.one.legend = T)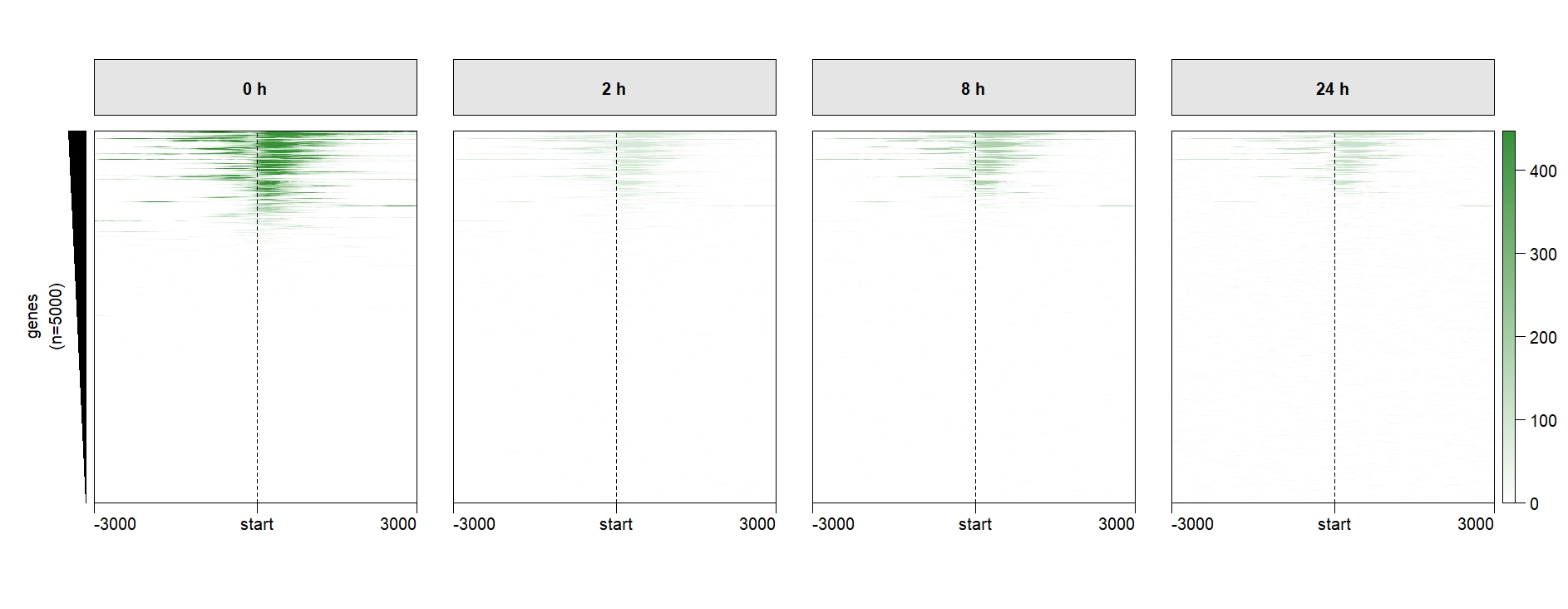
8.2 Row split
Split the rows:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
heatmap.col = col,
row.split = row_split)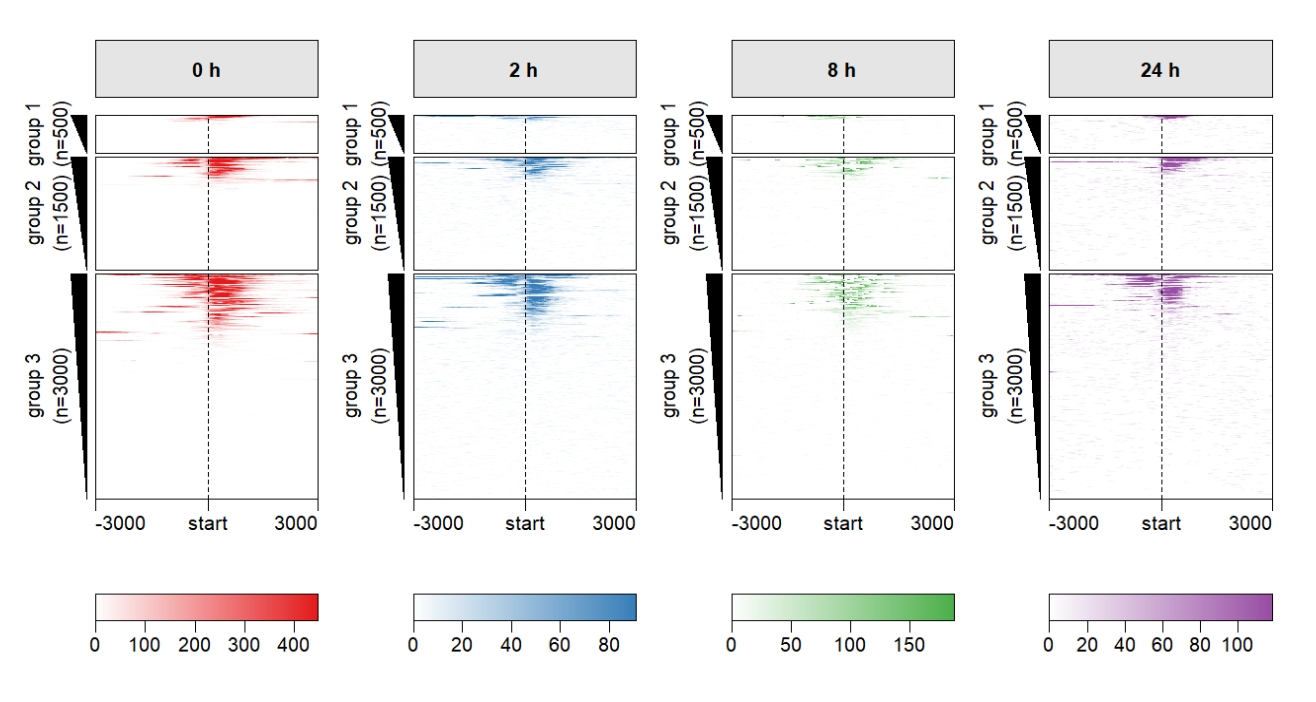 Keep only one legend:
Keep only one legend:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
row.split = row_split,
keep.one.left.annobar = T,
scale.range = T,
keep.one.legend = T)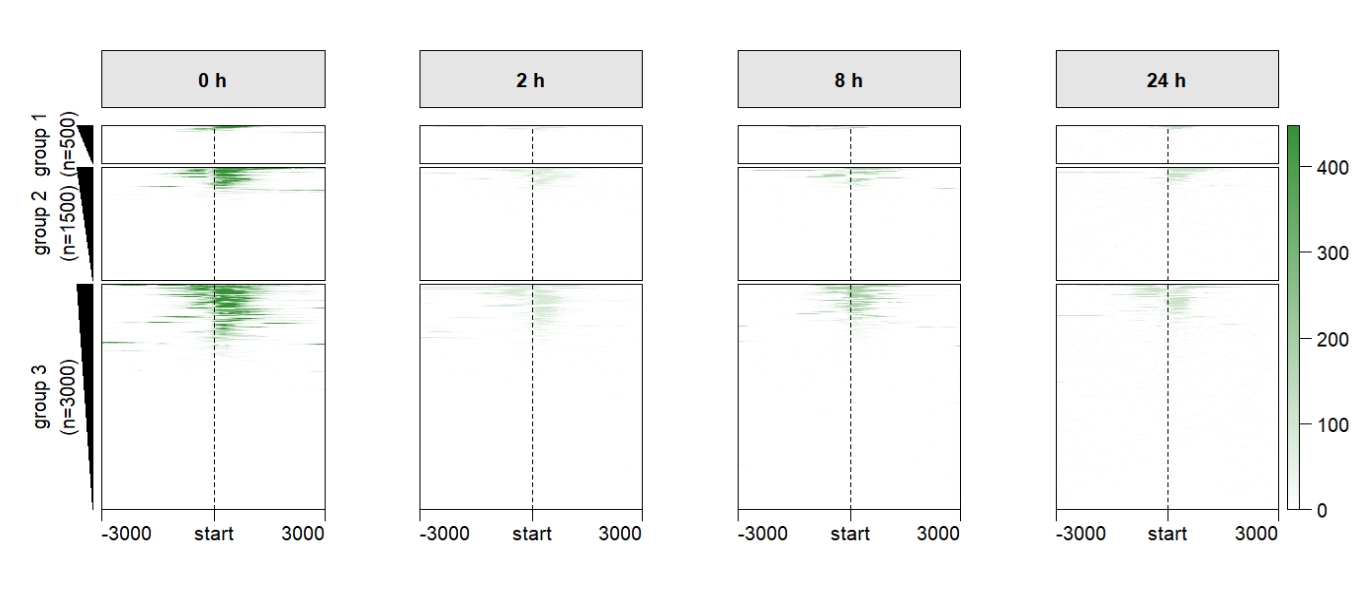 ## Draw profile
## Draw profile
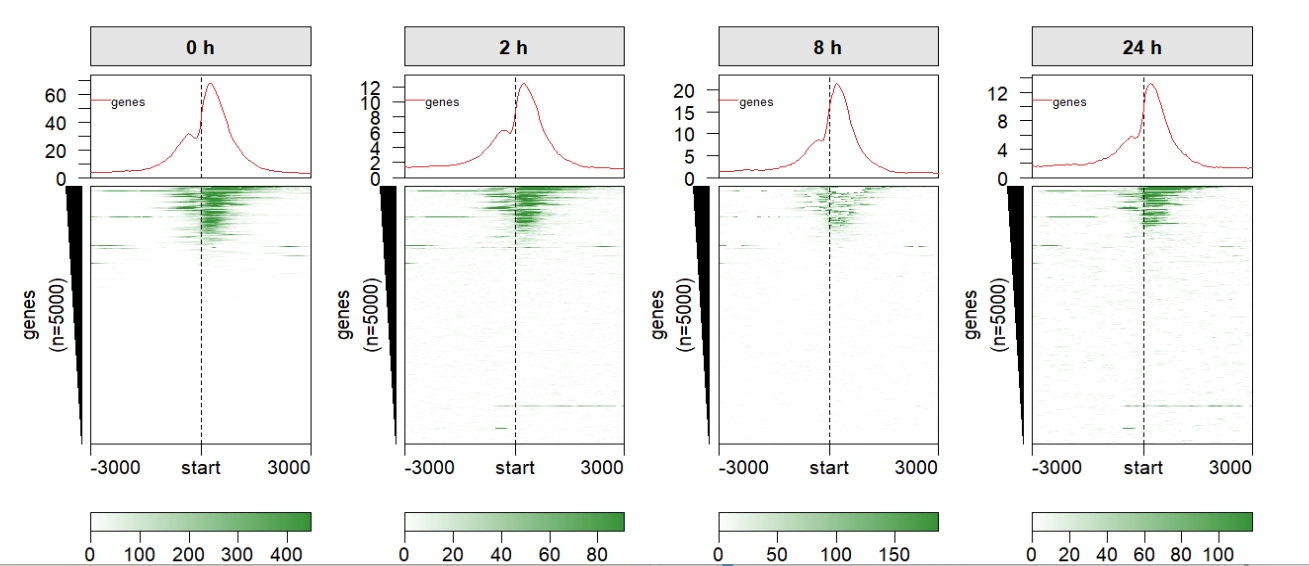
scale.y.range can make the multiple profile y scales to be same:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
draw.profile = T,
scale.y.range = T)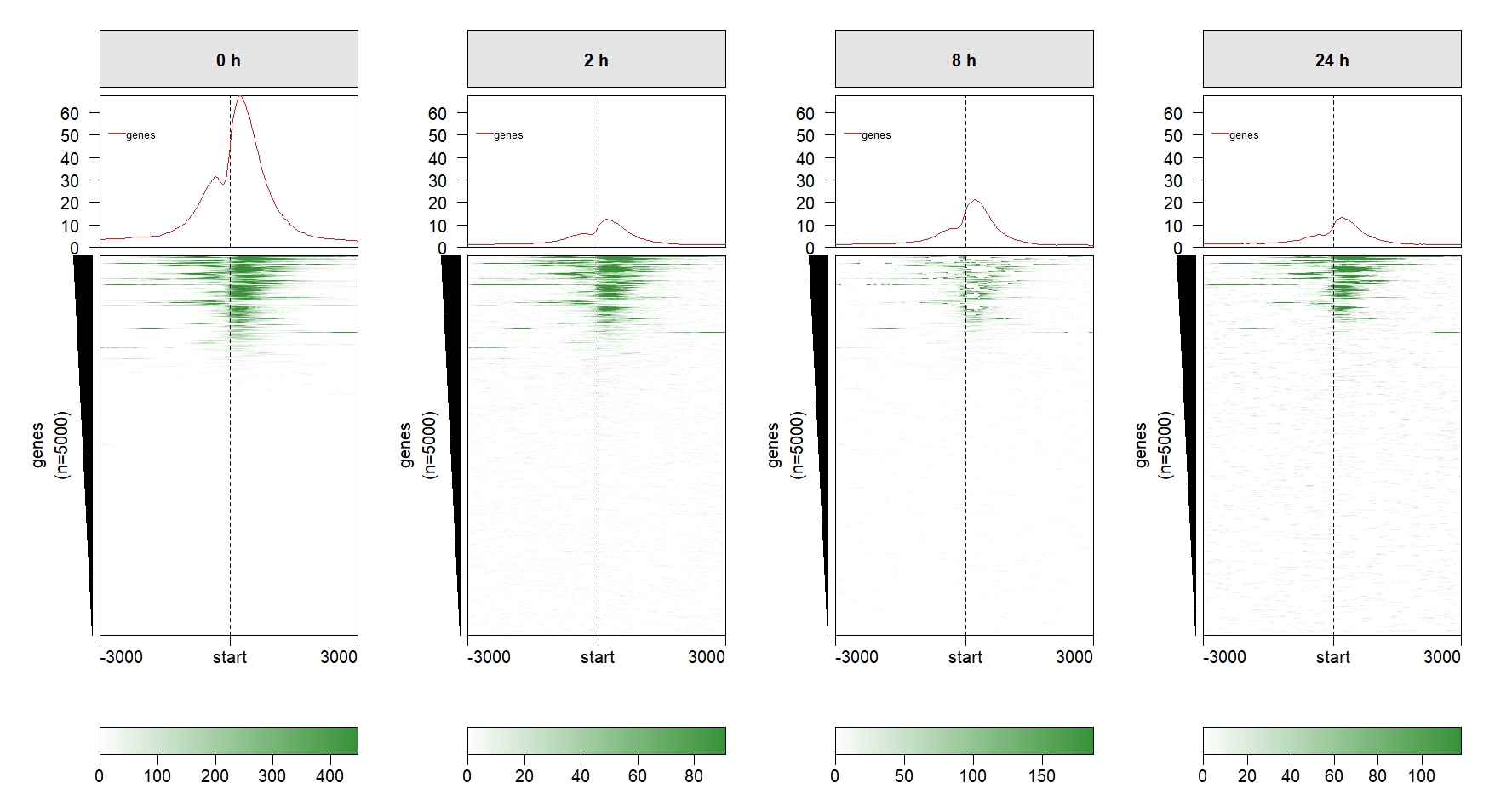 We can remove some redundant elements:
We can remove some redundant elements:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
draw.profile = T,
scale.y.range = T,
scale.range = T,
ChipHeatmap.params = list(HeatmapLayout.params = list(heatmap.size = c(0.9,0.5))),
keep.one.left.annobar = T,
keep.one.legend = T,
keep.one.line.legend = T,
keep.one.profile.yaxis = T)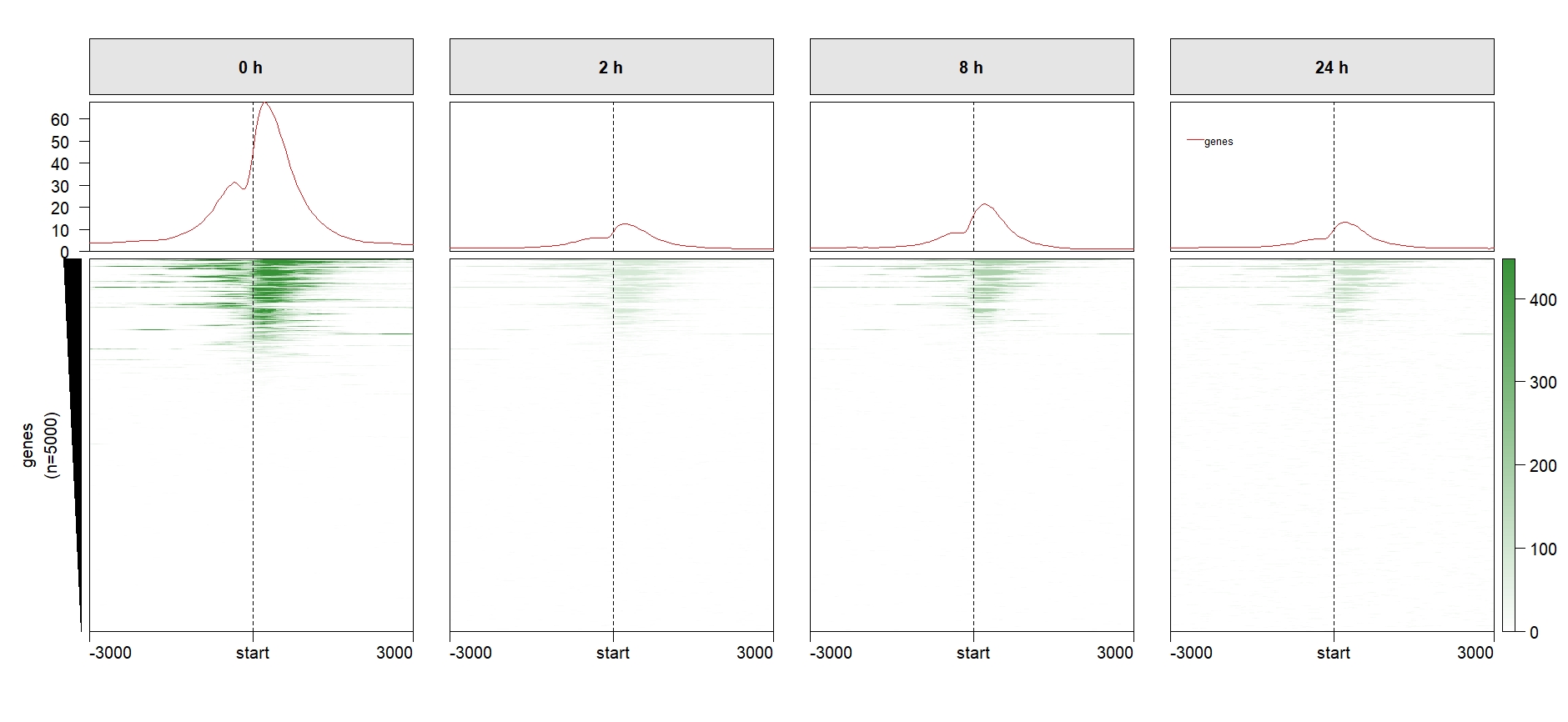 Add row split:
Add row split:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
draw.profile = T,
scale.y.range = T,
ChipHeatmap.params = list(HeatmapLayout.params = list(heatmap.size = c(1,0.5))),
plot.size = c(0.8,0.8),
scale.range = T,
keep.one.left.annobar = T,
keep.one.legend = T,
keep.one.line.legend = T,
keep.one.profile.yaxis = T,
row.split = row_split)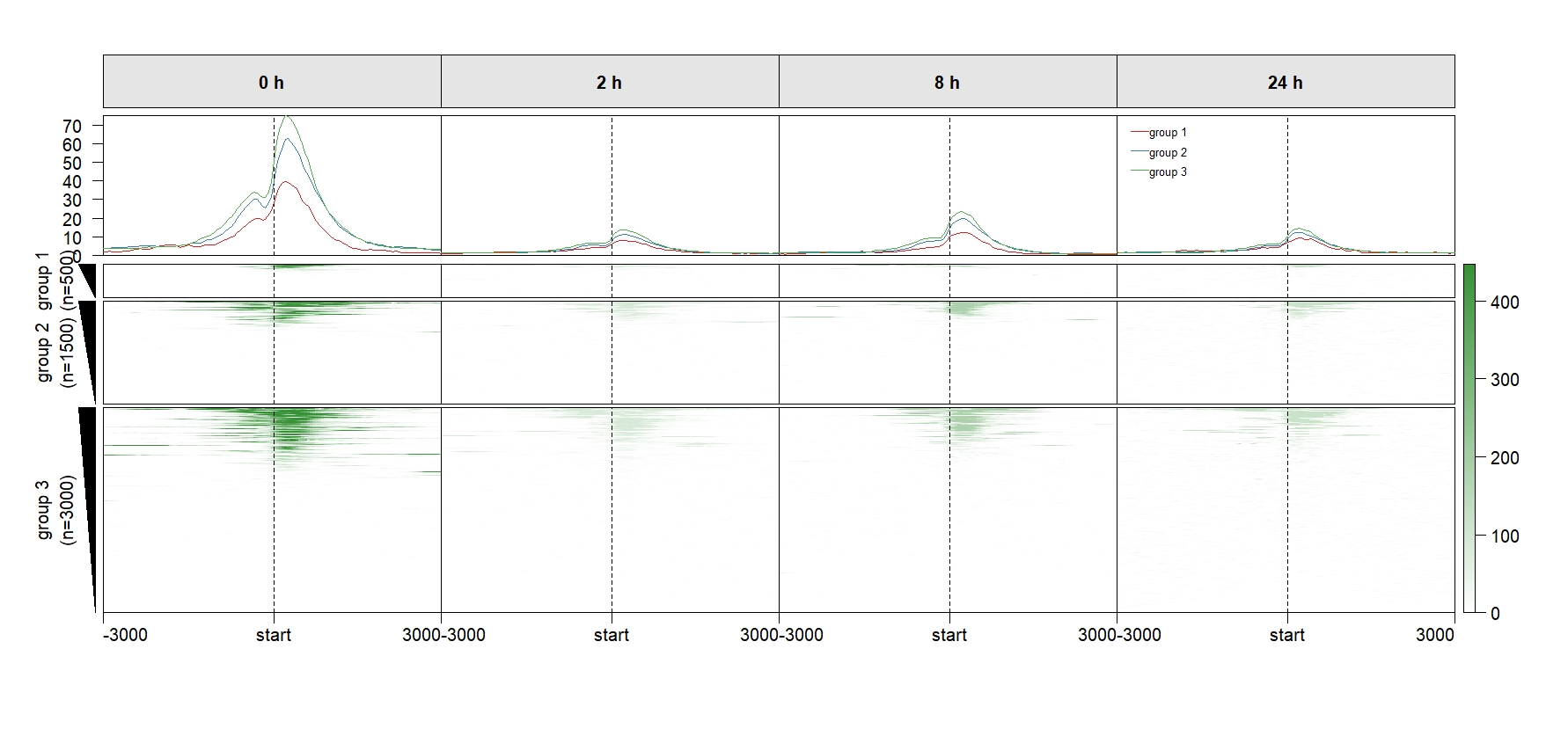
8.3 Add group for samples
You can supply a list to define the how the samples be groupyed by. You must keep the sample orders to be same with you group orders:
Add one group:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
sample.group1 = list(group1 = c("0 h","2 h"),
group2 = c("8 h","24 h")))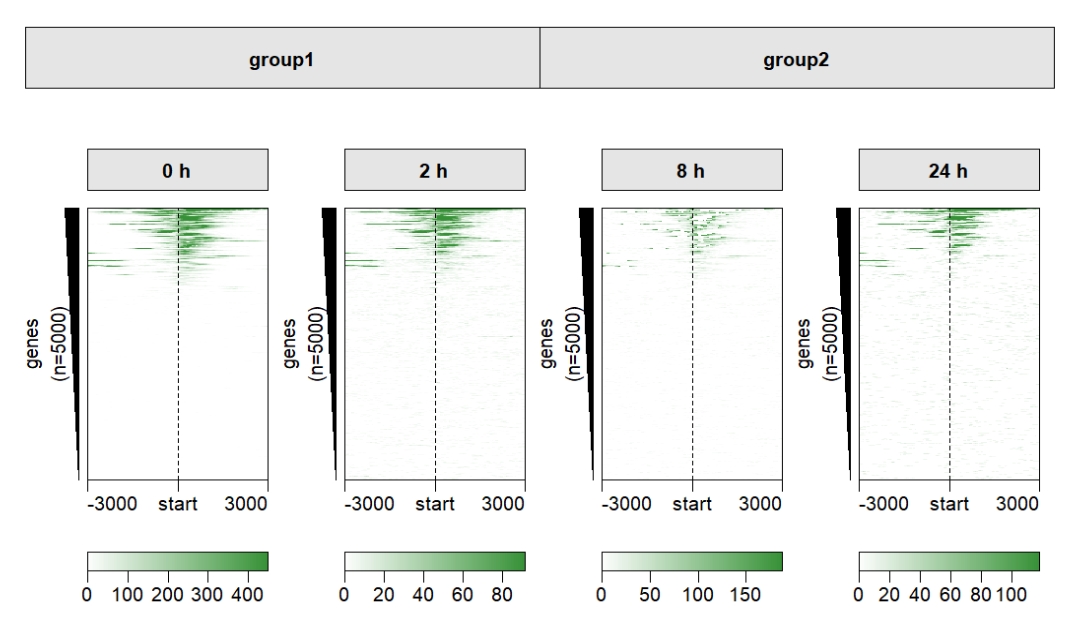
Add two groups:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
sample.group1 = list(group1 = c("0 h","2 h"),
group2 = c("8 h","24 h")),
sample.group2 = list(control = c("0 h","2 h","8h"),
treat = c("24 h")))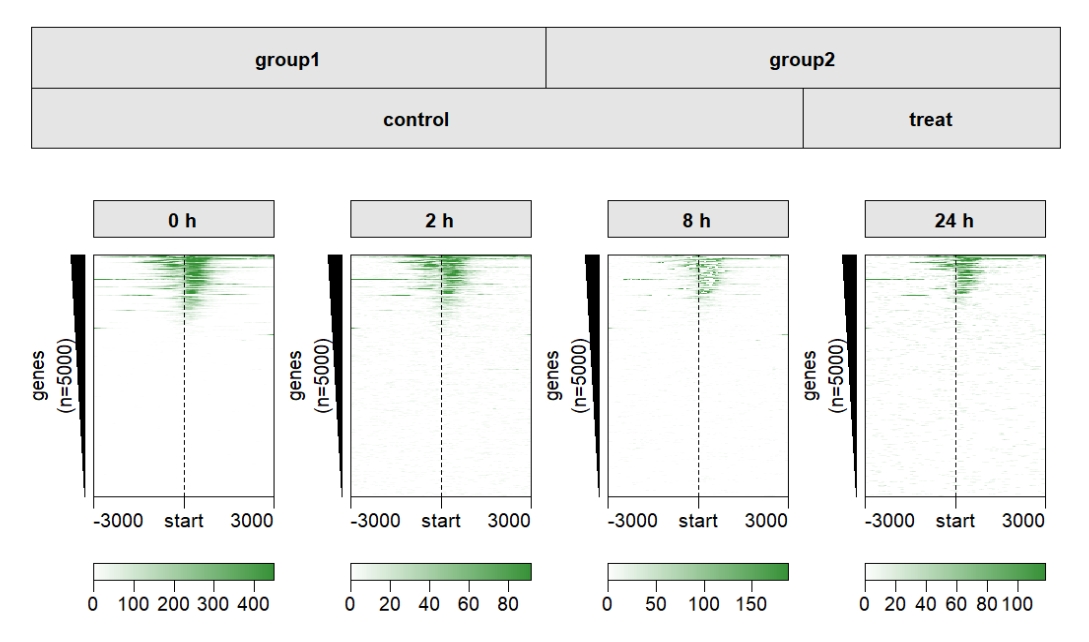
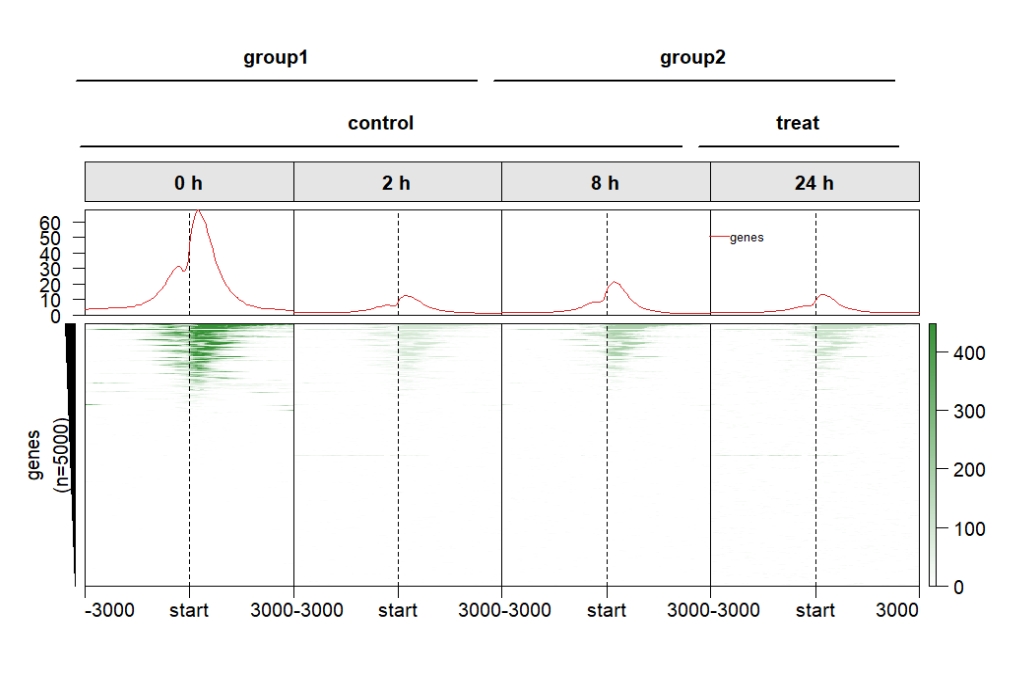
Control the top annobar style:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
sample.group1 = list(group1 = c("0 h","2 h"),
group2 = c("8 h","24 h")),
sample.group2 = list(control = c("0 h","2 h","8h"),
treat = c("24 h")),
draw.anno.fun.params.g1 = list(group.anno.rect.fill = c("orange","pink"),
group.anno.rect.col = NA),
draw.anno.fun.params.g2 = list(group.anno.rect.fill = c("purple","grey"),
group.anno.rect.col = NA))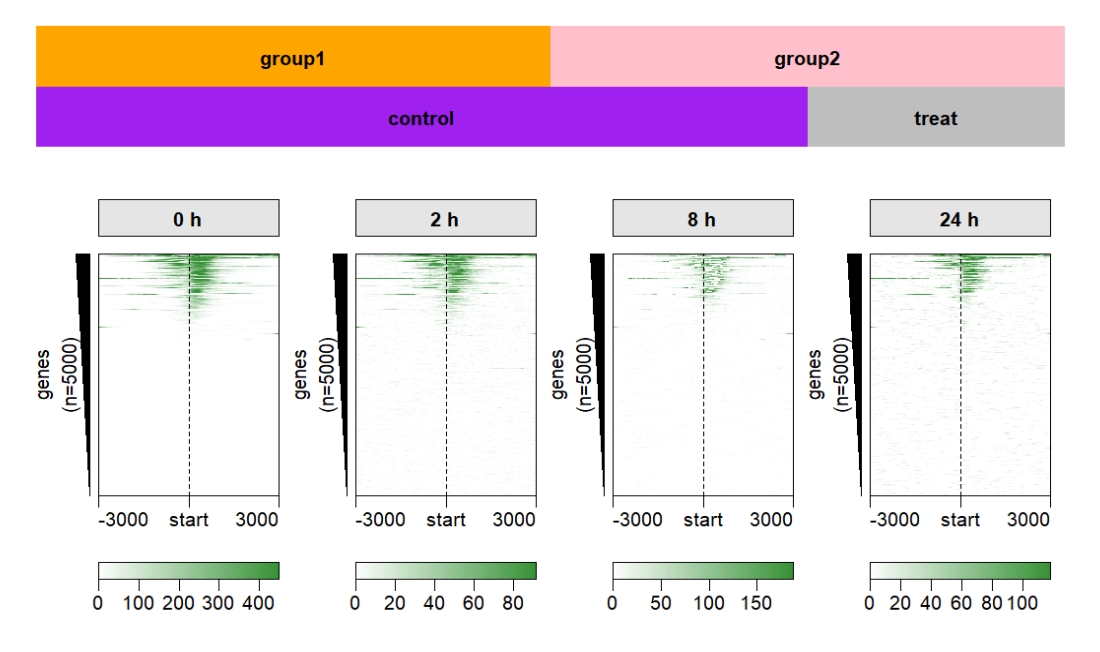 Control the top annobar height:
Control the top annobar height:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
sample.group1 = list(group1 = c("0 h","2 h"),
group2 = c("8 h","24 h")),
sample.group2 = list(control = c("0 h","2 h","8h"),
treat = c("24 h")),
draw.anno.fun.params.g1 = list(group.anno.rect.fill = c("orange","pink")),
draw.anno.fun.params.g2 = list(group.anno.rect.fill = c("purple","grey")),
panel.rect.gp = gpar(fill = "grey90"),
anno.panel.height = 0.05)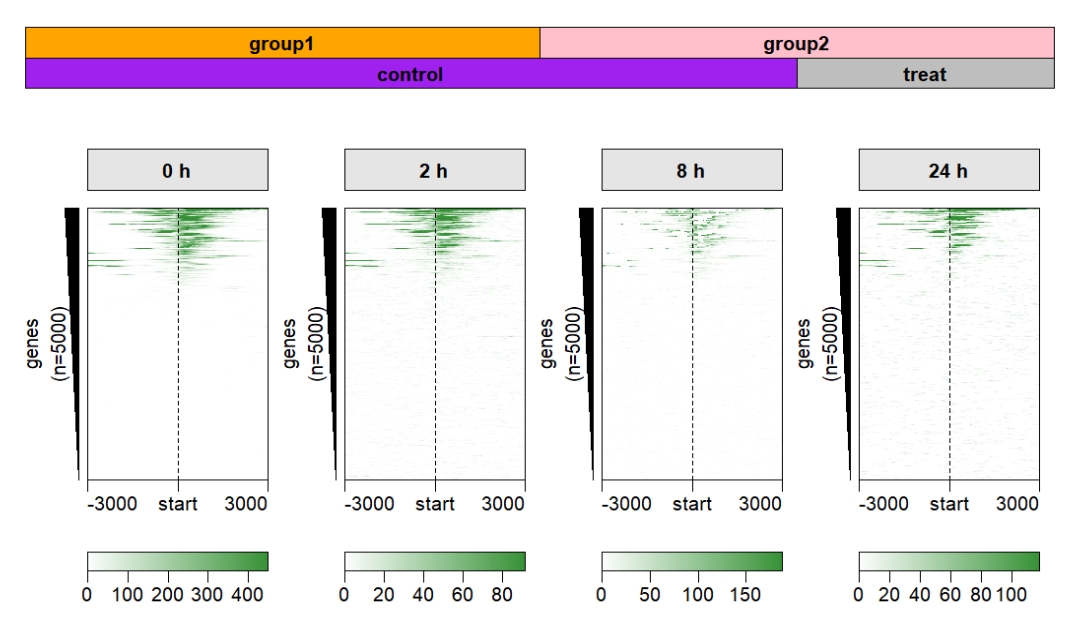 Add line anno:
Add line anno:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
sample.group1 = list(group1 = c("0 h","2 h"),
group2 = c("8 h","24 h")),
sample.group2 = list(control = c("0 h","2 h","8h"),
treat = c("24 h")),
draw.anno.fun.params.g1 = list(sample.anno.type = "line"),
draw.anno.fun.params.g2 = list(sample.anno.type = "line"))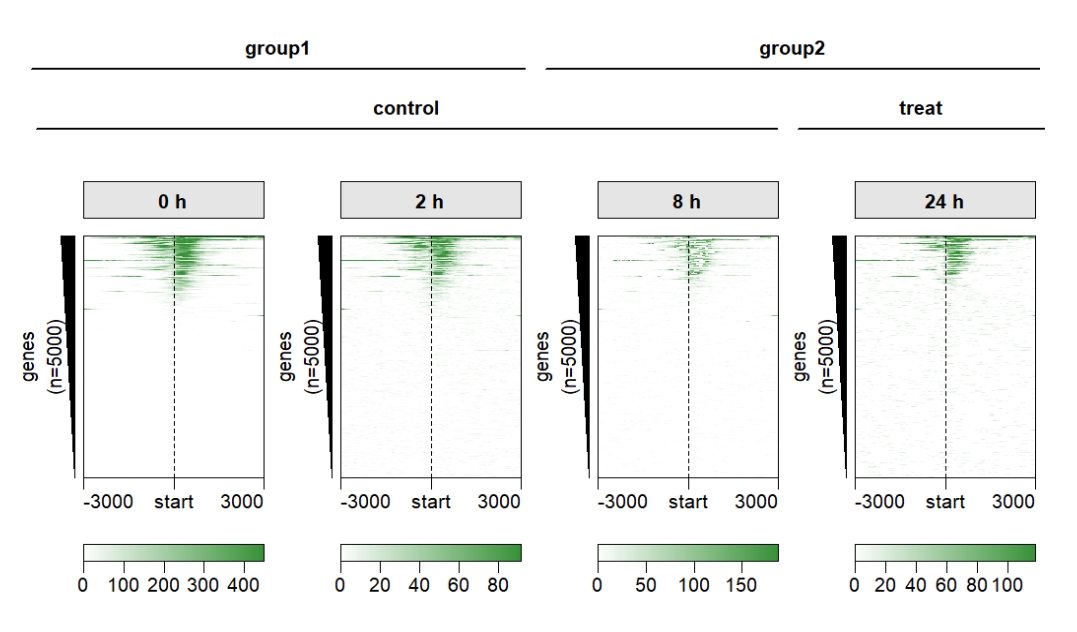 Draw a complete plot:
Draw a complete plot:
multiHeatmap(mat.list = deep_mat,
sample.label = c("0 h","2 h","8 h","24 h"),
sample.group1 = list(group1 = c("0 h","2 h"),
group2 = c("8 h","24 h")),
sample.group2 = list(control = c("0 h","2 h","8h"),
treat = c("24 h")),
draw.anno.fun.params.g1 = list(sample.anno.type = "line"),
draw.anno.fun.params.g2 = list(sample.anno.type = "line"),
scale.range = T,
scale.y.range = T,
draw.profile = T,
keep.row.order = T,
keep.one.left.annobar = T,
keep.one.profile.yaxis = T,
keep.one.line.legend = T,
keep.one.legend = T,
plot.size = c(0.75,0.9),
ChipHeatmap.params = list(HeatmapLayout.params = list(heatmap.size = c(1,0.5))))