8 Track_plot supplement
Here show some examples for track_plot function.
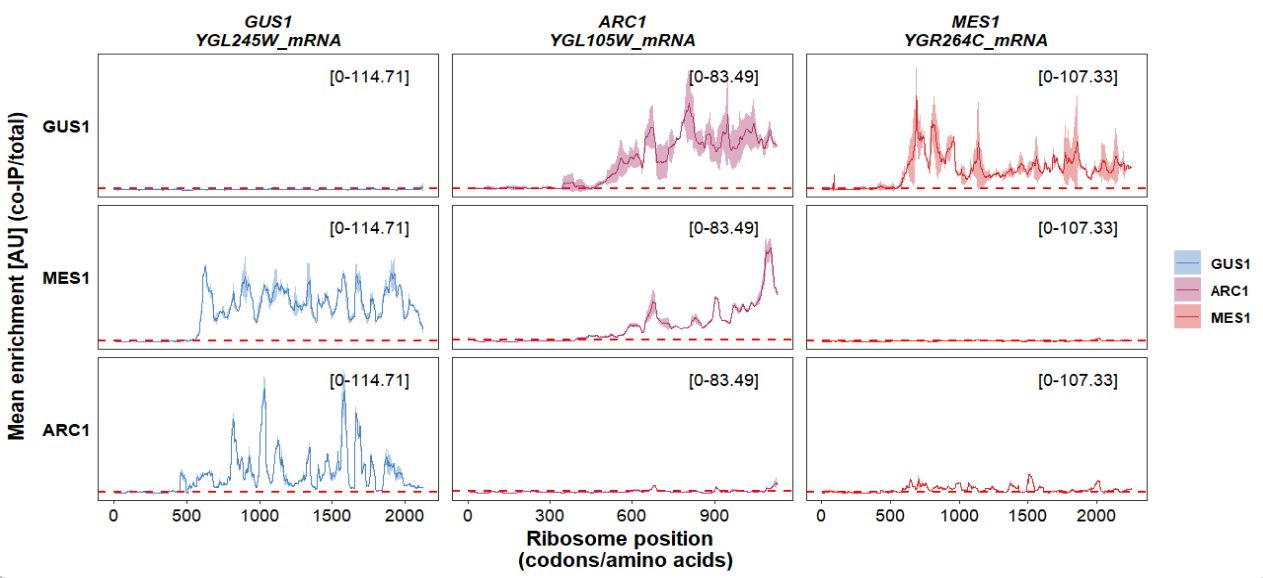
A simple plot:
track_plot(plot_type = "interactome",
signal_data = df_sw,
gene_order = c('GUS1','ARC1','MES1'),
sample_order = c('GUS1','MES1','ARC1'),
line_col = c('GUS1' = '#4883C6','ARC1' = '#AA356A','MES1' = '#D52C30'))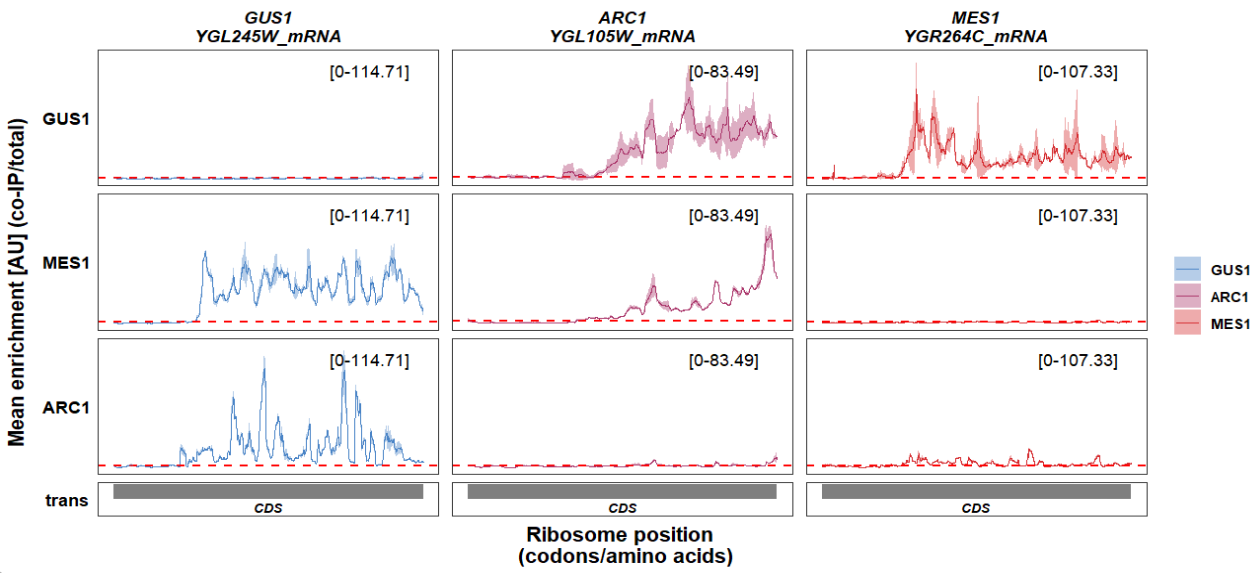
Remove trans panel border:
track_plot(plot_type = "interactome",
signal_data = df_sw,
gene_order = c('GUS1','ARC1','MES1'),
sample_order = c('GUS1','MES1','ARC1'),
line_col = c('GUS1' = '#4883C6','ARC1' = '#AA356A','MES1' = '#D52C30'),
remove_trans_panel_border = T)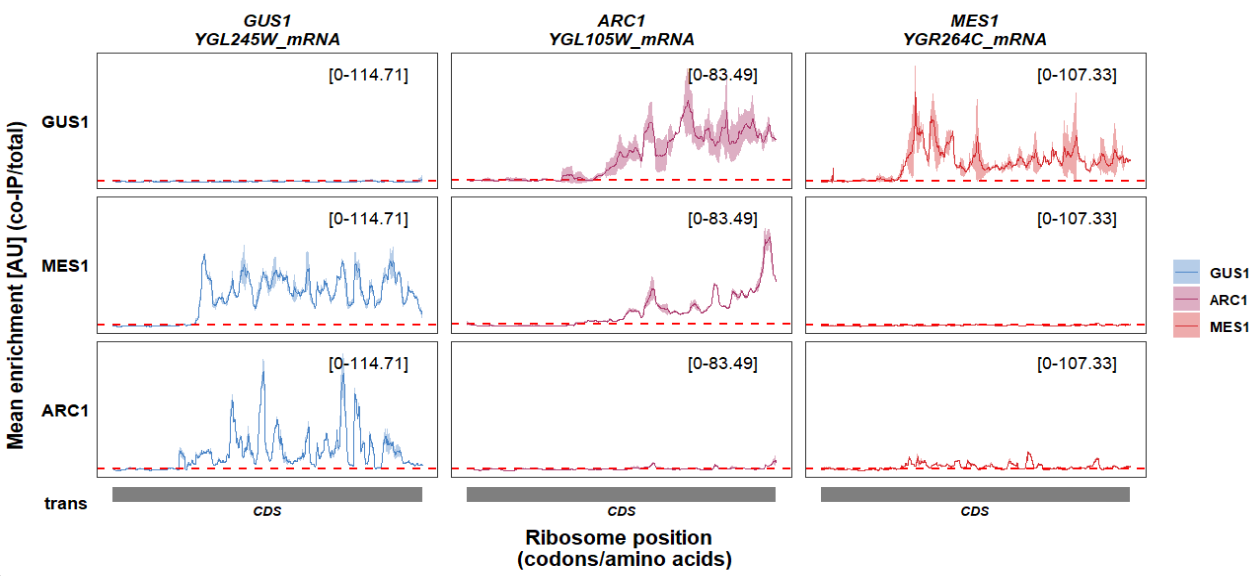
You can also remove all panel borders:
track_plot(plot_type = "interactome",
signal_data = df_sw,
gene_order = c('GUS1','ARC1','MES1'),
sample_order = c('GUS1','MES1','ARC1'),
line_col = c('GUS1' = '#4883C6','ARC1' = '#AA356A','MES1' = '#D52C30'),
remove_all_panel_border = T)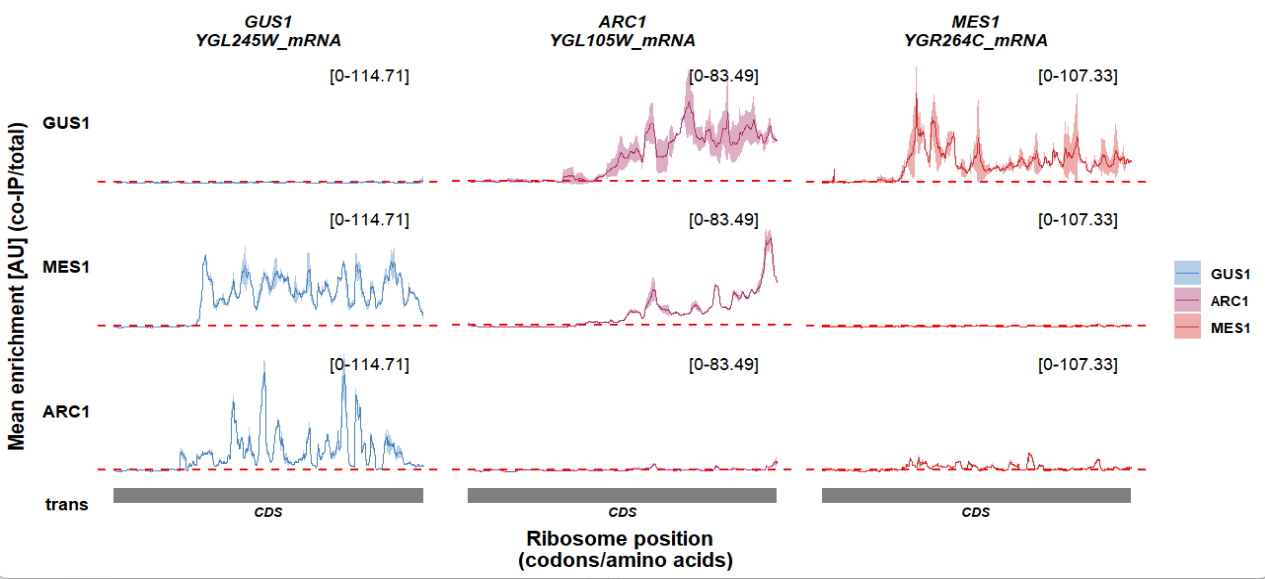
Show y axis ticks instead of text range:
track_plot(plot_type = "interactome",
signal_data = df_sw,
gene_order = c('GUS1','ARC1','MES1'),
sample_order = c('GUS1','MES1','ARC1'),
line_col = c('GUS1' = '#4883C6','ARC1' = '#AA356A','MES1' = '#D52C30'),
remove_trans_panel_border = T,
show_y_ticks = T)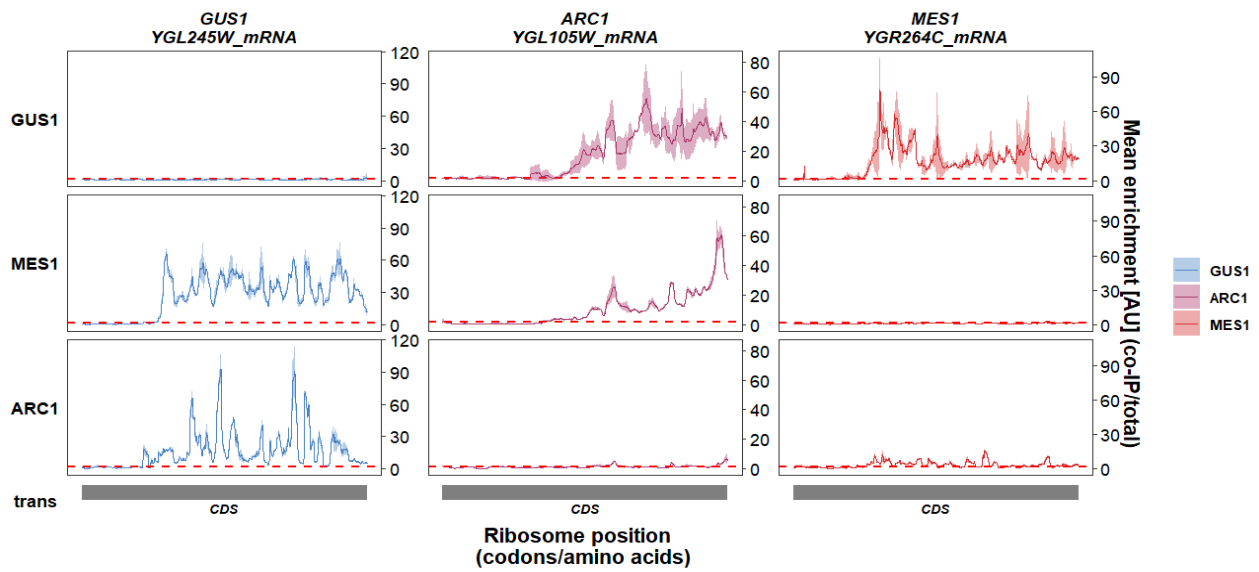
Modify details for multiple graph layers:
track_plot(plot_type = "interactome",
signal_data = df_sw,
gene_order = c('GUS1','ARC1','MES1'),
sample_order = c('GUS1','MES1','ARC1'),
line_col = c('GUS1' = '#4883C6','ARC1' = '#AA356A','MES1' = '#D52C30'),
remove_trans_panel_border = T,
geom_line_params = list(size = 1),
geom_hline_params = list(yintercept = 5,color = "black"),
geom_ribbon_params = list(alpha = 0.6))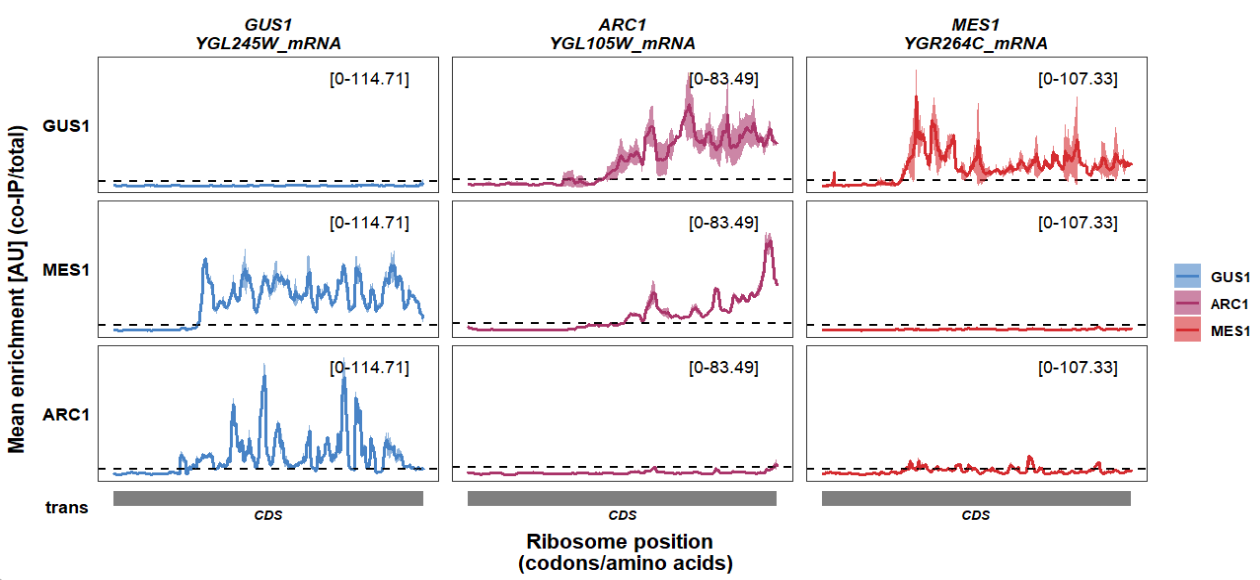
Turn off fixed y axis range by column:
track_plot(plot_type = "interactome",
signal_data = df_sw,
gene_order = c('GUS1','ARC1','MES1'),
sample_order = c('GUS1','MES1','ARC1'),
line_col = c('GUS1' = '#4883C6','ARC1' = '#AA356A','MES1' = '#D52C30'),
remove_trans_panel_border = T,
show_y_ticks = T,
fixed_col_range = F)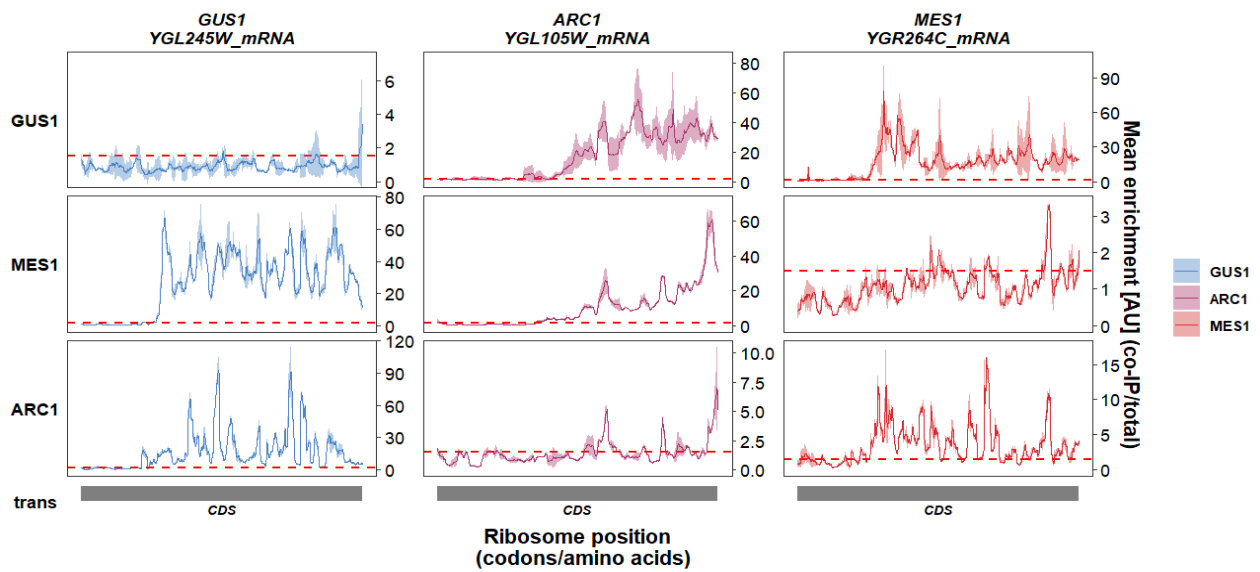
Show x axis:
track_plot(plot_type = "interactome",
signal_data = df_sw,
gene_order = c('GUS1','ARC1','MES1'),
sample_order = c('GUS1','MES1','ARC1'),
line_col = c('GUS1' = '#4883C6','ARC1' = '#AA356A','MES1' = '#D52C30'),
show_x_ticks = T)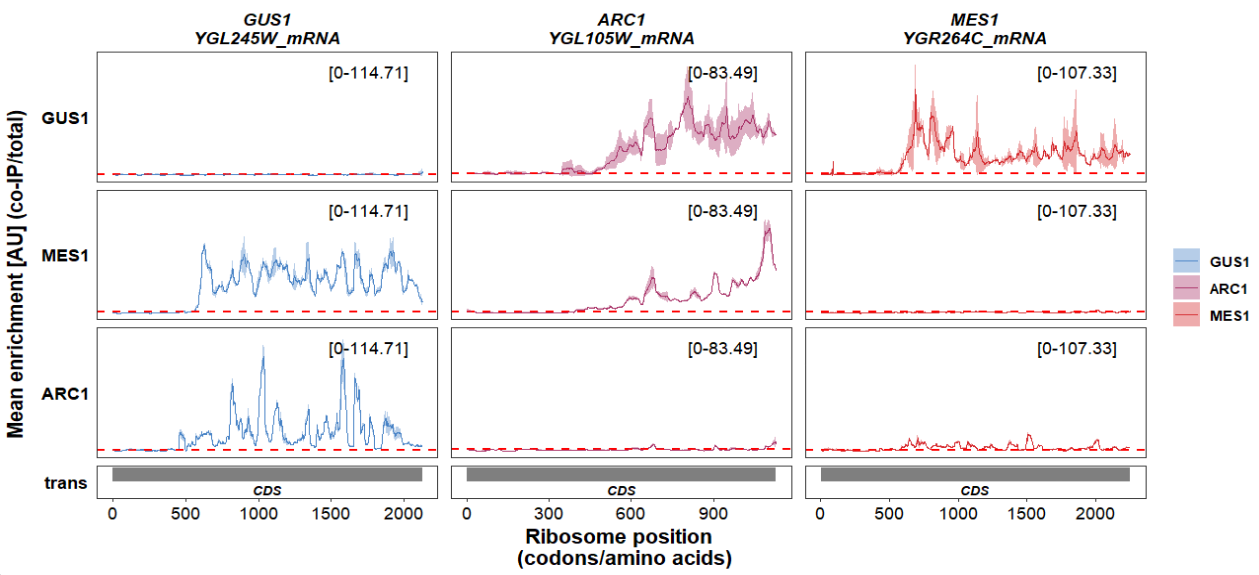
Remove gene structure panel:
track_plot(plot_type = "interactome",
signal_data = df_sw,
gene_order = c('GUS1','ARC1','MES1'),
sample_order = c('GUS1','MES1','ARC1'),
line_col = c('GUS1' = '#4883C6','ARC1' = '#AA356A','MES1' = '#D52C30'),
show_x_ticks = T,
add_gene_struc = F)