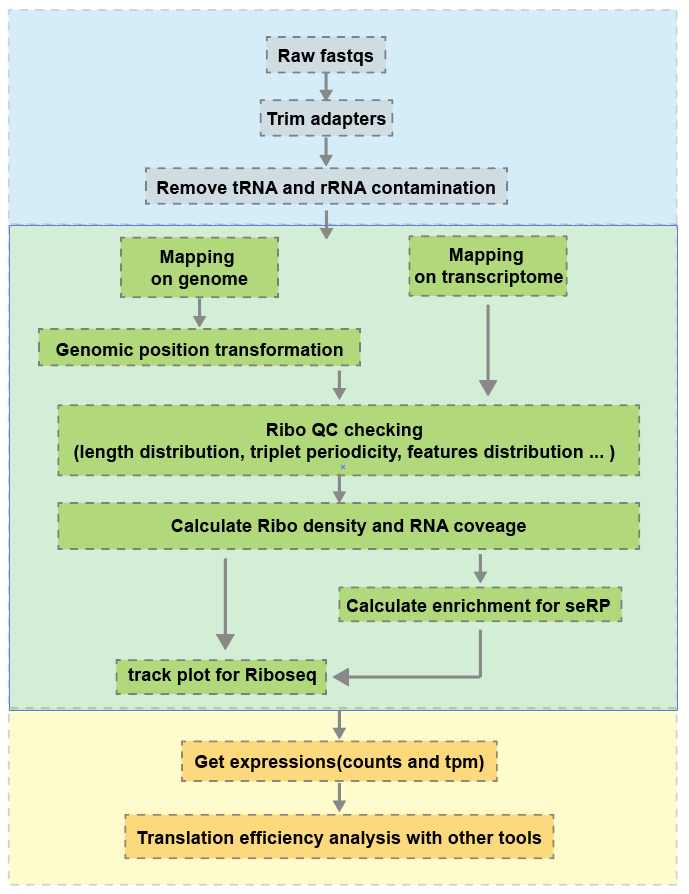
2 Workflow
RiboProfiler has two main mapping strategies: genome-based and transcriptome-based. Since mature RNA is typically bound by ribosomes for translation into protein, I recommend aligning reads to the transcriptome. If you choose to align reads to the genome instead, you can transform the resulting alignment positions to transcriptomic positions using the gene annotation file (GTF).
For transcriptome-based mapping, we select the longest CDS transcript as input. If multiple transcripts have the same CDS length, we choose the one with the longest exon length.
For paired end sequencing, just use one of them to perform the analysis such as read1.
Here’s a simplified flowchart to illustrate the process: